The real-time reverse transcription polymerase chain reaction – treat with caution
Posted: 21 September 2007 | | No comments yet
The real-time reverse transcription polymerase chain reaction (RT-qPCR) has become the enabling technology par excellence in every field of molecular research and development, including that of clinical drug development and discovery. Its ability to detect as well as quantify RNA biomarkers sensitively, specifically and speedily has made it an indispensable tool in translating the identification of complex biological processes into an understanding of their roles in disease pathology, response to therapy and associated pharmacological proof-of-concept drug efficacy and toxicity studies.
The real-time reverse transcription polymerase chain reaction (RT-qPCR) has become the enabling technology par excellence in every field of molecular research and development, including that of clinical drug development and discovery. Its ability to detect as well as quantify RNA biomarkers sensitively, specifically and speedily has made it an indispensable tool in translating the identification of complex biological processes into an understanding of their roles in disease pathology, response to therapy and associated pharmacological proof-of-concept drug efficacy and toxicity studies.
The real-time reverse transcription polymerase chain reaction (RT-qPCR) has become the enabling technology par excellence in every field of molecular research and development, including that of clinical drug development and discovery. Its ability to detect as well as quantify RNA biomarkers sensitively, specifically and speedily has made it an indispensable tool in translating the identification of complex biological processes into an understanding of their roles in disease pathology, response to therapy and associated pharmacological proof-of-concept drug efficacy and toxicity studies.
The principal advantages of the real-time polymerase chain reaction (qPCR) are its capacity to generate quantitative data over a wide dynamic range, coupled with high through-put, speed and convenience1, along with its potential for generating more reliable data2. The addition of a reverse transcription (RT) step3 has extended these benefits to the quantification of messenger, regulatory and genomic RNAs, making RT-qPCR to-day’s de facto standard for RNA analysis4,5, even in resource-limited settings6. RT-qPCR assays combine exquisite sensitivity and specificity; turn-around time from sample receipt to result can be less than two hours (the details of the assay5 and appropriate protocols7,8 are described elsewhere). They are performed in a closed system, thus minimising the risk of contamination and quantitative results are calculated using the threshold cycle (Ct), which is the cycle fraction when the real-time PCR instrument first detects the fluorescence generated during a successful amplification reaction. The Ct is obtained during the exponential part of the amplification process, when the most accurate measurement of accumulation is possible. The higher the starting copy number of the nucleic acid target, the sooner a significant increase in fluorescence is observed. Real-time monitoring permits not only accurate quantification of target copy numbers but also critical assessment of the reaction itself. The extensive choice of chemistries, enzymes and instrument platforms available for RT-qPCR is another reason for its immense appeal, albeit at the risk of substantial potential for the generation of discordant results9.
Applications
The technology has two main applications. Firstly, the characterisation of cellular RNA expression signatures, frequently following global expression profiling using microarrays10. Secondly, diagnostic evaluations such as detecting RNA viruses11, disease monitoring12, molecular staging13, drug regimen efficacy assays14 and many others15-17. The utility of RT-qPCR in quantifying cellular mRNA has been widely described18. A more recent enhancement has been its use for the analysis of micro RNA (miRNA) expression, which represents a level of post-transcriptional gene regulation that is not yet fully characterised. The probable role of miRNAs in cell differentiation, developmental biology and disease states19, coupled with their wide variation in abundance makes them ideal targets for RT-qPCR assays, as its wide dynamic range allows it to both detect extremely low expression and accurately quantify highly abundant miRNAs. RT-qPCR assays have been developed so that they can discriminate between the miRNA precursors and the biologically active mature molecules; they can even discriminate between two miRNAs that differ by as little as a single nucleotide20-22. Similarly, RT-qPCR offers an accurate and scalable method for validating and quantifying gene knockdown in RNAi experiments23.
RT-qPCR is widely used in pharmacological investigations: there are studies assessing the mechanisms of pharmacology and toxicity24-26, evaluating the pharmacokinetics and pharmacodynamics of dosing regimens27, measuring the kinetics of gene expression in response to specific drugs28,29 and evaluating the effects of diet on intestinal drug uptake30.
Alternatives
It is worth bearing in mind that there may not be a need for a real-time assay at all. For example, the qualitative detection of the presence of a genetically manipulated organism, of a pathogen, or a tumour cell may in itself be sufficient, allowing the use of a fluorescence-based end-point assay. Even if accurate quantification is necessary, other assay technologies may be more appropriate. LATE-PCR, a modified form of asymmetric PCR that efficiently generates single-stranded amplicons31, appears to be more sensitive for the small amounts of target material characteristic of single cell analysis carried out for pre-implantation diagnosis32. Alternatively, StaRT-PCR33 allows quantitative and quality-controlled multi-gene transcript abundance measurements and is widely used for the development of drugs, biomarkers and molecular diagnostics34,35. It may be more appropriate if standardisation and inter-laboratory reproducibility is a paramount requirement. Finally, there are non-PCR-based alternatives to PCR-based alternatives, for example isothermal methods such as the helicase-dependent DNA amplification36. These are not as efficient and robust as real-time PCR, but as they do not require complex thermal cyclers they may identify a niche in field-based applications such as public health and environmental science, or even point of care diagnostics37.
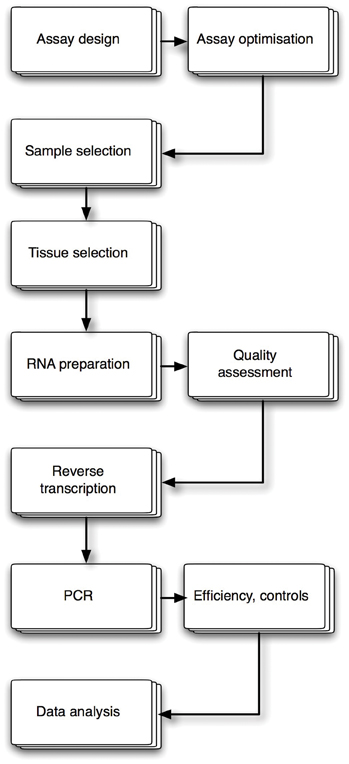
The assay design
Methodical assay design, combined with meticulous optimisation using step-by-step adjustments to protocols, reagents and conditions design, is the key to a successful RT-qPCR experiment (Figure 1). The establishment of standard operating procedures (SOP) is an essential requirement, especially if the assay is used for diagnostic purposes.

The real-time reverse transcription polymerase chain reaction (RT-qPCR) has become the enabling technology par excellence in every field of molecular research and development, including that of clinical drug development and discovery. Its ability to detect as well as quantify RNA biomarkers sensitively, specifically and speedily has made it an indispensable tool in translating the identification of complex biological processes into an understanding of their roles in disease pathology, response to therapy and associated pharmacological proof-of-concept drug efficacy and toxicity studies.
Ascertaining the purpose of the assay is another fundamental prerequisite, as this defines its mandatory performance metrics. These include delineation of the assay’s linearity, reproducibility and limits of sensitivity and specificity; whether a qualitative assay is sufficient or whether it needs to be quantitative, whether this is best achieved using one or several targets, either in separate reactions or as multiplexed assays; whether a probe-based or non-probe-based chemistry should be used, whether the RT-step should use specific or random priming and whether the assay should be a one enzyme/one tube, two enzyme/one tube or a two enzyme/two tube assay. A minimum of three replicate wells is required for the generation of confidence intervals, but in practice it is sufficient to run duplicate reactions. The use of single wells gives no information on well-to-well reproducibility. A standard curve with a minimum of four or five standards should be included to produce an accurate set of data. The standard curve must cover the dynamic range of the assay and should also be included with every run. Inclusion of a standard curve also provides an instant and consistent positive control.
Template
Standard procedures for sample acquisition and preparation are essential for reliable assay performance and it is becoming clear that stringent quality assessment of RNA is an essential necessity for reliable mRNA quantification8. Conventional means of assessing RNA quality, for example, gel-based visual inspection, OD260/280 ratios and even Agilent Bioanalzer RNAChip analyses are not sufficient, as they do not provide information about the integrity of mRNA itself. Instead, new approaches to assessing mRNA integrity are needed. One such method is the 3’:5’ assay that measures the integrity of one or more reference mRNAs8. Following oligo-dT-primed cDNA synthesis, two or three amplicons located at the 5’, centre and 3’ ends of the mRNA(s) are quantified in a multiplex reaction. Intact mRNA will result in 3’:5’ ratios of between 1 and 5, whereas degraded mRNA will result in 3’:5’ ratios of 50 or more. A separate quality control assay has been developed to detect the presence of reaction inhibitors38. A potato-specific target is amplified in the presence of RNA extracted from the samples of interest; in the absence of inhibition, copy numbers for the potato target are the same as those recorded for water controls. In the presence of inhibitors, copy numbers are reduced. These kinds of controls are the preferred means of quality assessing RNA samples and should be included in every SOP. One fact that is not always appreciated is that in real-life RNA, extraction efficiencies are variable, tissue-dependent and can be very low, which can result in recovery rates as low as 11% reported39.
The RT reaction
The cDNA synthesis step represents another critical factor, as results depend on the choice of priming strategy40 as well as on the type of reverse transcriptase used41. In principle, target-specific primers are preferable as the most sensitive means of priming cDNA42. However, this is not always so, and priming efficiency is target-gene dependent (Figure 2). When using specific RT primers, it is important to consider the structure of the target region, as intra-strand RNA complementarity will favour the re-annealing of double-stranded RNA, rather than hybridisation of the DNA primer to the RNA.
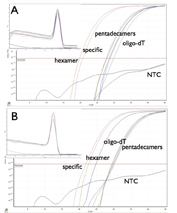

Figure 2: Target-dependent variation in priming efficiency. Typical SYBR Green results. A. Amplification plots for osteopontin showing the lowest Ct obtained from a reaction primed by specific primers, with hexamers more efficient than oligo-dT, which in turn is more efficient than various concentrations of pentadecamers. The melt curves show that all primers generate the same amplicon. B. Amplification plots for p21 showing the lowest Ct obtained from a reaction primed by hexamers, with specific primers more efficient than various concentrations of pentadecamers, which in turn are more efficient than oligo-dT. The melt curves show that all primers generate the same amplicon.
PCR
The PCR step itself is the most robust, and least troublesome segment of the RT-qPCR assay. The most important parameter here is the optimisation of the PCR efficiency, as small changes in efficiency will result in huge differences in calculated RNA copy numbers. Therefore, careful design of PCR primers is a crucial issue, as secondary structure and sequence composition within the PCR amplicon affect the efficiency, and hence the sensitivity of the assay43, and small differences in the forward and reverse primer ratio significantly affect the final result (Figure 3). Databases are now available with fluorogenic probe and primer sequences for numerous genes in humans, mice, rats and other species44-46. However, it is still essential to validate and optimise primers, even when using these assays.


Figure 3: Assay sensitivity depends on primer optimisation. The same forward and reverse primers were added to PCR reactions at concentrations of 50, 100 and 150 nM, respectively, in all nine possible ratios. The maximum difference in calculated copy numbers for the target differs by 50-fold.
Chemistries
The simplest and least expensive entry into real-time technology makes use of intercalating dyes such as SYBR Green to report the progress of the PCR reaction. Normally this simply requires the addition of the dye to an optimised conventional RT-PCR assay, hence conversion from legacy assays can be quickly implemented. DNA-binding dyes can even be used for multiplexing, for example using multiplexed tandem PCR (MT-PCR)47. This imaginative technique uses multiple gene-specific primer pairs, typically 30+, which are added to a RNA preparation from fresh or an archival sample, together with RT and Taq DNA polymerase. Following the RT step, the multiplexed cDNAs are simultaneously amplified for 10-15 cycles, distributed into multiple individual PCR reactions and single targets are amplified using primers nested inside the primers used for the multiplexed pre-amplification. MT-PCR can be configured for as little as 10 pg RNA; the amount of RNA typically contained within a single mammalian cell.
However, the minimalism that makes the use of intercalating dyes so attractive is also the source of substantial drawbacks; cDNA synthesis conditions48, primer dimers49 and even sequence composition50 can interfere with accurate quantification and require re-optimisation of assays. Furthermore, since DNA-binding dyes are non-specific, any PCR product, including primer dimers, will generate a signal. This requires the application of additional steps to ensure that only the amplification of genuine PCR products is being recorded. One approach includes dissociation, or melt curve, analysis, which can distinguish primer dimers from genuine amplification products. This involves the continuous monitoring of fluorescence as the temperature is slowly increased. Fluorescence decreases slowly with increasing temperature until the melting temperature characteristic of the specific amplicon is reached, at which time there is a more rapid decrease in the fluorescence. The point of maximum rate of change defines the melting temperature of the amplicon and plotting the rate of change in fluorescence, for example, the first derivative, verses temperature, identifies this point. Sometimes, however, it will become necessary to re-design primers and assay optimisation may become so laborious as to make a migration to a probe-based assay more productive.
The use of amplicon-specific fluorogenic probes corrects the many shortcomings of various non-probe-based chemistries. There are several such chemistries available, with the dual-label TaqMan probes by far the most popular51,52. These are linear probes containing two dyes, one of which quenches the fluorescent signal of a second reporter dye when they are in close molecular proximity to one another. During the PCR reaction, the fluorescent probe is cleaved by the Taq polymerase, thus separating the dyes and resulting in increased fluorescence with each amplification cycle. “Molecular Beacons” are an alternative design that adopt a hairpin structure while free in solution, bringing the fluorescent dye and quencher into close proximity53. When a beacon hybridises to a target, the fluorescent dye and quencher are separated and the fluorescent dye emits light upon irradiation. Unlike dual labelled probes, molecular beacons remain intact during the amplification reaction. Different probes can be labelled using different reporter dyes, thus enabling the simultaneous detection of several targets within a single reaction tube (multiplex assay). It is worth noting that the use of probe-based assays does not obviate the need to optimise primers. Although primer dimers are not detected, they will interfere with the efficiency of the assay and will contribute to poor assay performance. Many other chemistries are available and are discussed in detail elsewhere7.
Analysis
Data analysis and reporting remain subjective and contentious concerns. It is essential to adjust (‘normalise’) the raw data for differences in mRNA quality and concentrations, RT efficiencies and other experimental errors54. Normalisation can be carried out in a number of ways and there is no single answer as to which method is the most appropriate to use. However, internal standards are arguably the most appropriate procedure for cellular RNA quantification, and the current standard requires the normalisation of data against a minimum of three reference genes55. Certainly, the use of inappropriate reference genes will generate unreliable data56-58.
The question of what constitutes a negative no-template control (NTC) is not as clear-cut as might be imagined. A probe-based assay should always return negative NTCs and any sign of amplification must result in a repeat of the assay, as contamination is the most likely reason for the positive signal. In rare cases it is possible to obtain a positive signal due to baseline drift, where a degrading probe releases its fluorescence and the cumulative release of reporter from the quencher eventually results in fluorescence crossing the detection threshold. However, inspection of amplification plots will easily allow this to be determined. The situation is not as clear-cut when analysing data obtained using SYBR Green chemistry. Clearly, if the NTC is positive, and the Ct is near the Cts recorded for the test samples, the assay is suspect. However, if the melting curve reveals that the signal is generated by an amplicon with significantly lower Tm, it is most likely that the positive signal is caused by an artifact of primer dimers forming, encouraged by the absence of template. A related issue concerns the question of what constitutes a negative result for a test sample. To be confidently scored as negative, there should be no amplification detectable from the sample, there should also be no amplification from the NTCs but there must be a positive signal in the positive controls. Even then, the range of the positive controls only allows the statement that there was no target down to the limit of detection of the assay.
Instrumentation
Lastly, it is important to match the requirements of the assay with the capabilities of the qPCR instrument. In principle, all instruments can run and report data for a basic assay, with obvious differences only in terms of software user-friendliness and multiplexing potential. However, there are some key differences between instrument platforms and that must be factored into the assay design as they will have an impact on assay performance and can determine how robust and reproducible an assay will be59. Although most instruments use either a 96-well or a 384-well format, technical specifications vary, and there are significant differences in the evenness of the heating and cooling, the rates of temperature change as well as temperature stability and uniformity across the heating block. The use of rotor-based instruments addresses these problems60, albeit at the cost of introducing a less familiar assay format. Although somewhat subjective, user-friendliness of the software used to set up the assay and its reporting and analysis features also determine the suitability of an instrument for a particular type of assay. It is worth mentioning here that the more a company describes its software as user-friendly, the more cautious one needs to tread.
There have been interesting moves towards developing more portable thermal cycler technology61,62, which may be of use in field-based applications.
Final caveat
The main constraints of RT-qPCR, which at worst generates results that are open to inconsistent interpretation, have already been addressed. It should be clear that meaningful quantification depends on a rigorous application of standard operating procedures and transparent data interpretation. There is one other important caveat: the presence of an mRNA or miRNA, or changes in its abundance, is by itself not biologically significant unless variation is accompanied by matching changes in protein amount or activity8. Consequently, quantification of RNA cannot simply be extrapolated to pronouncements on protein abundance or activity. This makes any biological interpretation of quantitative mRNA data challenging and should lead to more cautious conclusions with regards to the relevance of RT-qPCR data.
The future
Expression analysis technology continues to advance in leaps and bounds, exemplified by Illumina’s digital gene expression tag profiling that can generate global signatures through sequencing without requiring any sequence knowledge. Whilst technically impressive, this is a sledgehammer approach that fails to take note of the fact that many investigations require the analysis of fewer and fewer numbers of cells. This is because tissue biopsies are made up of numerous different cell types, each with their own individual expression profiles. This is most obvious in cancer tissue, where the cancer itself is heterogeneous and it is surrounded by heterogeneous normal tissue. It means that reliance on global expression signatures obtained from large amounts of tissue or cells can mask significant differences in individual expression patterns, which are likely to provide critical information about intercellular signalling and regulation.
Given the limited amounts of target often available, it is necessary to be able to increase the information that can be obtained from individual samples. Traditionally, RT-qPCR assays have been used to study a very limited number of targets in a large number of samples. Multiplexing is one way of maximising information from a small sample, but it is difficult to develop complex multiplexing protocols, as there is a limit to the number of dyes that are available and there can be loss of sensitivity as well as a reduction in the dynamic range of the overall assay. Crucially, conventional 96-well, rotor and even 384-well -based RT-qPCR assays require relatively significant amounts of template, making the analysis of multiple targets from a single sample without pre-amplification difficult.
Consequently, the progress of miniaturisation microfluidic technologies that allow massively parallel PCR to be carried out in combination with a thousand-fold reduction in the reaction volume, is an exciting development63. They will also make assays affordable to be run, thus allowing the inclusion of more controls. Indeed, the power to carry out digital PCR64,65 may replace the need for real-time PCR altogether. Digital PCR involves carrying out PCR analyses on extremely dilute samples partitioned into thousands of individual assays. Successful amplification in any one partition reflects the signal from a single template molecule. The proportion of positive amplifications among the total number of PCR reactions analysed allows the calculation of template concentration in the original undiluted sample. Because template molecule quantification by digital PCR does not rely on dose- response relationships between reporter dyes and nucleic acid concentrations, its analytical precision should be superior to that of real-time PCR.
In conclusion, RT-qPCR technology is extremely powerful and can quantify RNA levels accurately, provided standard reaction conditions and analysis criteria are followed. For now it remains the method of choice, although it is clear that new technology developments allowing easy digital PCR may supersede RT-qPCR, at least for high through-put screening.
References
- Heid CA, Stevens J, Livak KJ and Williams PM (1996). Real time quantitative PCR. Genome Res. 1996; 6:986-994.
- Niedrig M, Meyer H, Panning M and Drosten C (2006). Follow-up on diagnostic proficiency of laboratories equipped to perform orthopoxvirus detection and quantification by PCR: the second international external quality assurance study. J Clin Microbiol 2006; 44:1283-1287.
- Gibson UE, Heid CA and Williams PM (1996). A novel method for real time quantitative RT-PCR. Genome Res. 1996; 6:995-1001.
- Bustin SA (2002). Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol 2002; 29:23-39.
- Bustin SA (2000). Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000; 25:169-193.
- Rouet F and Rouzioux C (2007). The measurement of HIV-1 viral load in resource-limited settings: how and where? Clin Lab 2007; 53:135-148.
- Bustin SA Editor (2004). A-Z of quantitative PCR. IUL Press, La Jolla, CA.
- Nolan T, Hands RE and Bustin SA (2006). Quantification of mRNA using real-time RT-PCR. Nat Protocols 2006; 1:1559-1582.
- Bustin SA (2005). Real-time, fluorescence-based quantitative PCR: a snapshot of current procedures and preferences. Expert Rev Mol Diagn 2005; 5:493-498.
- Provenzano M and Mocellin S (2007). Complementary techniques: validation of gene expression data by quantitative real time PCR. Adv Exp Med Biol 2007; 593:66-73.
- King DP, Ferris NP, Shaw AE, Reid SM, Hutchings GH, Giuffre AC, Robida JM, Callahan JD, Nelson WM and Beckham TR (2006). Detection of foot-and-mouth disease virus: comparative diagnostic sensitivity of two independent real-time reverse transcription-polymerase chain reaction assays. J Vet Diagn Invest 2006; 18:93-97.
- Lo-Coco F and Ammatuna E (2007). Front line clinical trials and minimal residual disease monitoring in acute promyelocytic leukemia. Curr Top Microbiol Immunol 2007; 313:145-156.
- Bustin SA (2006). Nucleic acid quantification and disease outcome prediction in colorectal cancer. Personalized Medicine 2006; 3:207-216.
- Kofla G and Ruhnke M (2007). Development of a new real-time TaqMan PCR assay for quantitative analyses of Candida albicans resistance genes expression. J Microbiol Methods 2007; 68:178-183.
- Bustin SA and Mueller R (2005). Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin Sci (Lond) 2005; 109:365-379.
- Anderson S, Howard B, Hobbs GR and Bishop CP (2005). A method for determining the age of a bloodstain. Forensic Sci Int 2005; 148:37-45.
- Chen ZW, Liu YY, Wu JF, She Q, Jiang CY and Liu SJ (2007). Novel bacterial sulfur oxygenase reductases from bioreactors treating gold-bearing concentrates. Appl Microbiol Biotechnol 2007; 74:688-698.
- Bustin SA, Benes V, Nolan T and Pfaffl MW (2005). Quantitative real-time RT-PCR–a perspective. J Mol Endocrinol 2005; 34:597-601.
- Bartel DP (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-297.
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ and Guegler KJ (2005). Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 2005; 33:e179.
- Fu HJ, Zhu J, Yang M, Zhang ZY, Tie Y, Jiang H, Sun ZX and Zheng XF (2006). A novel method to monitor the expression of microRNAs. Mol Biotechnol 2006; 32:197-204.
- Tang F, Hajkova P, Barton SC, O’Carroll D, Lee C, Lao K and Surani MA (2006). 220-plex microRNA expression profile of a single cell. Nat Protocols 2006; 1:1154-1159.
- Tuzmen S, Kiefer J and Mousses S (2007). Validation of short interfering RNA knockdowns by quantitative real-time PCR. Methods Mol Biol 2007; 353:177-203.
- Guo Y, Jolly RA, Halstead BW, Baker TK, Stutz JP, Huffman M, Calley JN, West A, Gao H, Searfoss GH, Li S, Irizarry AR, Qian HR, Stevens JL and Ryan TP (2007). Underlying mechanisms of pharmacology and toxicity of a novel PPAR agonist revealed using rodent and canine hepatocytes. Toxicol Sci 2007; 96:294-309.
- Rokushima M, Omi K, Araki A, Kyokawa Y, Furukawa N, Itoh F, Imura K, Takeuchi K, Okada M, Kato I and Ishizaki J (2007). A toxicogenomic approach revealed hepatic gene expression changes mechanistically linked to drug-induced hemolytic anemia. Toxicol Sci 2007; 95:474-484.
- Liu J, Xie Y, Cooper R, Ducharme DM, Tennant R, Diwan BA and Waalkes MP (2007). Transplacental exposure to inorganic arsenic at a hepatocarcinogenic dose induces fetal gene expression changes in mice indicative of aberrant estrogen signaling and disrupted steroid metabolism. Toxicol Appl Pharmacol 2007; 220:284-291.
- Zhou Q, Guo P, Wang X, Nuthalapati S and Gallo JM (2007). Preclinical pharmacokinetic and pharmacodynamic evaluation of metronomic and conventional temozolomide dosing regimens. J Pharmacol Exp Ther 2007; 321:265-275.
- Rioja I, Bush KA, Buckton JB, Dickson MC and Life PF (2004). Joint cytokine quantification in two rodent arthritis models: kinetics of expression, correlation of mRNA and protein levels and response to prednisolone treatment. Clin Exp Immunol 2004; 137:65-73.
- Hu R, Hebbar V, Kim BR, Chen C, Winnik B, Buckley B, Soteropoulos P, Tolias P, Hart RP and Kong AN (2004). In vivo pharmacokinetics and regulation of gene expression profiles by isothiocyanate sulforaphane in the rat. J Pharmacol Exp Ther 2004; 310:263-271.
- Glaeser H, Bailey DG, Dresser GK, Gregor JC, Schwarz UI, McGrath JS, Jolicoeur E, Lee W, Leake BF, Tirona RG and Kim RB (2007). Intestinal drug transporter expression and the impact of grapefruit juice in humans. Clin Pharmacol Ther 2007; 81:362-370.
- Sanchez JA, Pierce KE, Rice JE and Wangh LJ (2004). Linear-after-the-exponential (LATE)-PCR: an advanced method of asymmetric PCR and its uses in quantitative real-time analysis. Proc Natl Acad Sci U S A 2004; 101:1933-1938.
- Hartshorn C, Anshelevich A and Wangh LJ (2005). Rapid, single-tube method for quantitative preparation and analysis of RNA and DNA in samples as small as one cell. BMC Biotechnol 2005; 5:2.
- Willey JC, Knight CR, Crawford EL, Olson DE, Hammersly J, Yoon Y and Sharief I (2004). Use of standardized reverse transcription-polymerase chain reaction and the standardized expression measurement center in multi-institutional trials to develop meaningful lung cancer classification based on molecular genetic criteria. Chest 2004; 125:155S-156S.
- Saccomanno CF (2006). Gene Express Inc. Pharmacogenomics 2006; 7:793-796.
- Peters EH, Rojas-Caro S, Brigell MG, Zahorchak RJ, des Etages SA, Ruppel PL, Knight CR, Austermiller B, Graham MC, Wowk S, Banks S, Madabusi LV, Turk P, Wilder D, Kempfer C, Osborn TW and Willey JC (2007). Quality-controlled measurement methods for quantification of variations in transcript abundance in whole blood samples from healthy volunteers. Clin Chem 2007; 53:1030-1037.
- Vincent M, Xu Y and Kong H (2004). Helicase-dependent isothermal DNA amplification. EMBO Rep 2004; 5:795-800.
- Gill P, Amini M, Ghaemi A, Shokouhizadeh L, Abdul-Tehrani H, Karami A and Gilak A (2007). Detection of Helicobacter pylori by enzyme-linked immunosorbent assay of thermophilic helicase-dependent isothermal DNA amplification. Diagn Microbiol Infect Dis 2007.
- Nolan T, Hands RE, Ogunkolade BW and Bustin SA (2006). SPUD: a qPCR assay for the detection of inhibitors in nucleic acid preparations. Anal Biochem 2006; 351:308-310.
- Lovatt A (2002). Applications of quantitative PCR in the biosafety and genetic stability assessment of biotechnology products. J Biotechnol. 2002; 82:279-300.
- Stahlberg A, Hakansson J, Xian X, Semb H and Kubista M (2004). Properties of the reverse transcription reaction in mRNA quantification. Clin Chem 2004; 50:509-515.
- Stahlberg A, Kubista M and Pfaffl M (2004). Comparison of reverse transcriptases in gene expression analysis. Clin Chem 2004; 50:1678-1680.
- Bustin SA and Nolan T (2004). Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech 2004; 15:155-166.
- Peters IR, Helps CR, Hall EJ and Day MJ (2004). Real-time RT-PCR: considerations for efficient and sensitive assay design. J Immunol Methods 2004; 286:203-217.
- Wang X and Seed B (2003). A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res 2003; 31:e154.
- Pattyn F, Speleman F, De Paepe A and Vandesompele J (2003). RTPrimerDB: the real-time PCR primer and probe database. Nucleic Acids Res 2003; 31:122-123.
- Pattyn F, Robbrecht P, De Paepe A, Speleman F and Vandesompele J (2006). RTPrimerDB: the real-time PCR primer and probe database, major update 2006. Nucleic Acids Res 2006; 34:D684-688.
- Stanley KK and Szewczuk E (2005). Multiplexed tandem PCR: gene profiling from small amounts of RNA using SYBR Green detection. Nucleic Acids Res 2005; 33:e180.
- Lekanne Deprez RH, Fijnvandraat AC, Ruijter JM and Moorman AF (2002). Sensitivity and accuracy of quantitative real-time polymerase chain reaction using SYBR green I depends on cDNA synthesis conditions. Anal.Biochem. 2002; 307:63-69.
- Vandesompele J, De Paepe A and Speleman F (2002). Elimination of primer-dimer artifacts and genomic coamplification using a two-step SYBR green I real-time RT-PCR. Anal.Biochem. 2002; 303:95-98.
- Giglio S, Monis PT and Saint CP (2003). Demonstration of preferential binding of SYBR Green I to specific DNA fragments in real-time multiplex PCR. Nucleic Acids Res 2003; 31:e136.
- Livak KJ, Flood SJ, Marmaro J, Giusti W and Deetz K (1995). Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR.Methods Appl. 1995; 4:357-362.
- Holland PM, Abramson RD, Watson R and Gelfand DH (1991). Detection of specific polymerase chain reaction product by utilizing the 5′—-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc.Natl.Acad.Sci.U.S.A. 1991; 88:7276-7280.
- Tyagi S and Kramer FR (1996). Molecular beacons: probes that fluoresce upon hybridization. Nat.Biotechnol. 1996; 14:303-308.
- Huggett J, Dheda K, Bustin S and Zumla A (2005). Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 2005; 6:279-284.
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A and Speleman F (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3:0034.0031-0034.0011.
- Tricarico C, Pinzani P, Bianchi S, Paglierani M, Distante V, Pazzagli M, Bustin SA and Orlando C (2002). Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal. Biochem. 2002; 309:293-300.
- Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G and Zumla A (2004). Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 2004; 37:112-119.
- Dheda K, Huggett JF, Chang JS, Kim LU, Bustin SA, Johnson MA, Rook GA and Zumla A (2005). The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem 2005; 344:141-143.
- Reynisson E, Josefsen MH, Krause M and Hoorfar J (2006). Evaluation of probe chemistries and platforms to improve the detection limit of real-time PCR. J Microbiol Methods 2006; 66:206-216.
- Herrmann MG, Durtschi JD, Bromley LK, Wittwer CT and Voelkerding KV (2006). Amplicon DNA melting analysis for mutation scanning and genotyping: cross-platform comparison of instruments and dyes. Clin Chem 2006; 52:494-503.
- Belgrader P, Young S, Yuan B, Primeau M, Christel LA, Pourahmadi F and Northrup MA (2001). A battery-powered notebook thermal cycler for rapid multiplex real-time PCR analysis. Anal.Chem. 2001; 73:286-289.
- Higgins JA, Nasarabadi S, Karns JS, Shelton DR, Cooper M, Gbakima A and Koopman RP (2003). A handheld real time thermal cycler for bacterial pathogen detection. Biosens Bioelectron 2003; 18:1115-1123.
- Morrison T, Hurley J, Garcia J, Yoder K, Katz A, Roberts D, Cho J, Kanigan T, Ilyin SE, Horowitz D, Dixon JM and Brenan CJ (2006). Nanoliter high throughput quantitative PCR. Nucleic Acids Res 2006; 34:e123.
- Pohl G and Shih Ie M (2004). Principle and applications of digital PCR. Expert Rev Mol Diagn 2004; 4:41-47.
- Ottesen EA, Hong JW, Quake SR and Leadbetter JR (2006). Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science 2006; 314:1464-1467.
Stephen A. Bustin
Academic Department of Surgery, Institute of Cell and Molecular Science, Queen Mary’s School of Medicine and Dentistry, University of London
Stephen Bustin obtained his PhD from Trinity College , University of Dublin in molecular genetics. His research interests centre around investigating the process of invasion and metastasis in colorectal cancer, in particular the role played by metastasis-associated oncogenes, signaling cascades and suppressor genes. His specific aim is to identify metastasis-associated expression signatures that will allow the inclusion of molecular parameters into clinical tumour staging, thus improving prognostic accuracy for individual cancer patients. The ultimate aim is the identification of specific sub-groups of patients that will benefit from new, individualised molecular therapeutic strategies. His laboratory continues to operate at the forefront of technological development in nucleic acid quantification. He is an internationally acknowledged leader in this area.




