Cancer immunotherapy using RNAi
Posted: 25 January 2007 | | 1 comment
Immunotherapy has recently emerged as an attractive form of treatment for cancer due to the potential of the immune system to eradicate tumours without inflicting damage on normal tissue. However, natural immune responses are usually inadequate to control cancer progression and require enhancement by vaccines.
Immunotherapy has recently emerged as an attractive form of treatment for cancer due to the potential of the immune system to eradicate tumours without inflicting damage on normal tissue. However, natural immune responses are usually inadequate to control cancer progression and require enhancement by vaccines.
Immunotherapy has recently emerged as an attractive form of treatment for cancer due to the potential of the immune system to eradicate tumours without inflicting damage on normal tissue. However, natural immune responses are usually inadequate to control cancer progression and require enhancement by vaccines.
These vaccines should aim to generate improved T cell-mediated immunity to tumour-associated antigens, as this type ofimmune response is crucial in the host defense against cancers. Such an aim may be achieved through innovative immunotherapeutic strategies that modify the properties of dendritic cells (DCs), the most potent activators of T cells. Recently, the attenuation of signaling pathways that negatively influence the stimulation of T cells by DCs has been explored as a potentially promising approach to cancer vaccination. Specifically, it may be of interest to suppress the expression of proteins produced by DCs which inhibit T cell priming, cause undesirable types of T helper cell responses, or induce apoptosis in the DCs themselves. RNA interference (RNAi) techniques have been shown to be an effective means of knocking down gene expression in cells, and may be successfully applied to modulate the properties of DCs for cancer immunotherapy. Nevertheless, several practical concerns need to be resolved prior to the implementation of RNAi technology in humans. Due to the swift progress of research in the biology of cancer, molecules and the immune system, it is likely that this technology may soon translate into a potent form of gene silencing in the clinical arena with profound applications to cancer immunotherapy.
Conventional treatments for cancer, such as surgery, chemotherapy and radiation therapy are frequently unable to cure late stage malignancies without causing severe side effects in patients. Thus, there is a great need for a novel cancer therapy that is capable of successfully treating these late stage malignancies. Over the past decade, immunotherapy has emerged as a promising alternative form of cancer treatment that may eradicate metastasised tumours without significantly damaging normal tissue. The host immune response is carried out by a variety of cell types that are capable of efficiently killing tumour cells when activated. Furthermore, the central components of the adaptive immune system, T and B cells, possess a diverse array of antigen receptors, which allows them to identify tumour cells in a highly specific manner. Therefore, cancer vaccines that effectively activate T and B cells to recognise tumour cells should facilitate the elimination of tumours at multiple sites of the body with minimal toxicity to surrounding tissue.
The synthesis of tumour-specific neutralising antibodies by B cells also constitutes an important arm of the antitumour immune response; however, the discussion in this article is limited exclusively to T cell-mediated immunotherapy. Among the cells of the immune system, T cells are primarily responsible for the control of cancers and therefore activation of these cells is crucial for generating antitumour immunity. CD8+ cytotoxic T lymphocytes (CTLs) are able to kill tumour cells either by secreting cytolytic granules or by activating death receptor-associated apoptosis pathways. By contrast, CD4+ T helper (Th) cells secrete cytokines that either inhibit tumour cell proliferation or assist in activating CTLs. Due to the critical role of T cells in mediating antitumour immune responses, cancer vaccine research has focused on the development of innovative strategies to enhance T cell activation and function.
It is well established that dendritic cells (DCs), the most potent antigen-presenting cells (APCs), are the strongest activators of T cells. In the major histocompatibility complex (MHC) class I (MHC-1)-restricted pathway of antigen presentation, cytoplasmic protein antigens are degraded into peptide fragments by the proteasome and loaded onto MHC-1 molecules for the priming of CD8+ T cells. Alternatively, DCs can phagocytose exogenous antigens into lysosomes and endosomes, where proteases, together with the acidic environment of these compartments, process the antigen for complexing with MHC class II (MHC-2) molecules and subsequent presentation to CD4+ T cells. The interactions between MHC molecules and T cell receptors, along with costimulatory signaling, causes T cells to undergo clonal expansion and differentiation. These activated T cells are capable of migrating to tumour tissue, where they either directly kill tumour cells (in the case of CTLs) or promote CTL proliferation and activation (in the case of Th cells). The understanding of the processes involved in DC-mediated activation of T cells has created many opportunities to enhance cancer immunotherapy by modifying the properties of DCs.
Several novel interventions that focus on attenuating the expression of specific proteins in DCs have been explored as a means for enhancing cancer immunotherapy. Vaccinations for cancer involve the delivery of tumour-associated antigens (TAAs) into DCs, resulting in the processing and presentation of these antigens, which leads to the generation of large populations of tumour-specific CTLs and Th cells that are capable of controlling tumours. Thus, modifying the properties of DCs represents an attractive method for enhancing T cell activation. These modifications may include the following:
- Inhibiting pathways that negatively regulate antigen presentation by DCs
- Reducing the expression of proteins that induce apoptosis in DCs
- Reducing the expression of proteins in DCs that lead to undesirable types of T cell-mediated immune responses
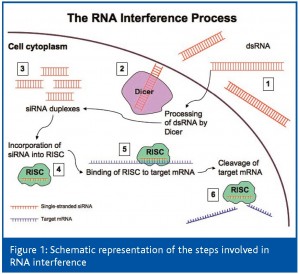
The suppression of these proteins that hinder effective immune responses to tumours should enable the improvement of cancer immunotherapy. RNA interference (RNAi) technology has recently emerged as a potent and specific method of gene silencing and may be used to regulate the expression of key proteins in DCs for the enhancement of tumour-specific immunity. This highly gene-specific phenomenon was first observed in the late 1980s and since then, its mechanism of action has become better understood1-3. Figure 1 illustrates the steps of the RNAi process. The first event in RNAi involves the cleavage of cytoplasmic double-stranded RNAs (dsRNA) into 21-28 nucleotide duplexes by the RNase-III enzyme Dicer. These fragments, referred to as small interfering RNAs (siRNA), are then incorporated into a multiprotein RNA-inducing silencing complex (RISC). The siRNA serves as a template for the recognition of complementary target mRNA, which is cleaved by the RISC enzyme Slicer (Argonaute-2) in the centre of the region homologous to the duplex, thus resulting in mRNA degradation and gene silencing. Alternatively, RISC binding can cause translational arrest of the target mRNA. Therefore, RNAi can serve both as a cellular defense (for instance, against viruses or transposons) and as a gene control mechanism in developmental processes. Also, RNAi displays relatively long-term efficacy because during Slicer activity, siRNA is protected within the RISC and is preserved to facilitate mRNA degradation in subsequent endonucleolytic reactions. Multiple types of small artificial dsRNA have been inserted into a variety of cells and organisms and have demonstrated that RNAi can be widely employed as a powerful tool for specific gene silencing.
dsRNA1 is delivered into the cell, where it is processed by the RNase-III enzyme Dicer2 into 21-28 nucleotide double-stranded siRNA duplexes3. One strand of the siRNA duplex is incorporated into RISC4 and acts as a template for the recognition of target mRNA5. The RISC enzyme Slicer cleaves the target mRNA and is preserved for further RNAi activity6.
This article discusses several strategies for cancer immunotherapy using RNAi technology to modify DCs. We also explore potential methods for the administration of RNAi in vivo as well as practical considerations for the clinical application of this technique. Altogether, we hope to show that RNAi technology provides a promising tool for cancer immunotherapy and may soon be feasible to implement in the clinical arena.
Modification of dendritic cells using RNAi technology for cancer immunotherapy
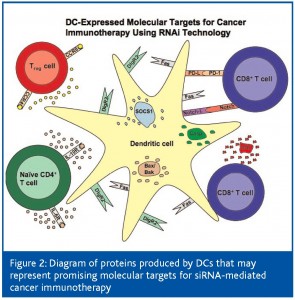
DCs possess high numbers of MHC-1 and MHC-2, costimulatory and intercellular adhesion molecules, and are therefore capable of activating T cells with remarkable efficiency. Additionally, DCs have the ability to migrate to secondary lymphoid organs where the majority of T cells are located. Thus, the modification of DCs to augment their capacity to prime naïve T cells represents a particularly effective strategy for the enhancement of the antitumour immune response. Current research efforts in this area have focused on inhibition of immunosuppressive proteins expressed by DCs, on preferential selection of a Th1 immune response and on prolongation of DC life. RNAi technology has been employed to silence genes in DCs that negatively influence antigen presentation and survival. These studies have resulted in the generation of elevated quantities of tumour-specific CTLs, which has been correlated with considerable antitumour effects. Thus, siRNA-mediated knockdown of immunosuppressive or proapoptotic proteins in DCs provides a novel and appealing approach for the enhancement of cancer immunotherapy. Figure 2 summarises the molecules produced by DCs that can potentially be targeted using RNAi technology to boost the antitumour immune response.
Inhibition of immunosuppressive proteins expressed by Dendritic Cells
DCs express a variety of immunosuppressive proteins that negatively influence T cell-mediated immunity, which can lead to inadequate immune responses to tumours. For example, DC-mediated activation of T cells is critically regulated by the suppressor of cytokine signaling 1 (SOCS1) protein. SOCS1 disrupts signaling pathways associated with interferon (IFN)-γ, interleukin (IL)-2, IL- 6, IL-7, IL-12, and IL-15 in T cells4 and is also believed to suppress antigen presentation to T cells5. Several studies have shown that administration of siRNA targeting SOCS1 into DCs loaded with tumour antigen leads to an increase in the number of tumour-specific T cells and inhibits the growth of established tumours in mice4,6,7. In addition, the immunosuppressive tryptophan- degrading enzyme indoleamine-2,3-dioxygenase (IDO) is produced in considerable amounts by plasmacytoid DCs8,9. Secretion of IDO by these DCs in the tumour-draining lymph nodes depletes tryptophan availability in the local microenvironment, which may inhibit the mitosis and function of nearby T cells 8 or induce profound anergy in these T cells10. Thus, knockdown of IDO or SOCS1 with siRNA represents an effective strategy that may be used to enhance cancer vaccines.
The presence of certain surface receptors on the DC membrane can inhibit T cell proliferation and may also provide excellent molecular targets for siRNA-mediated immunotherapy. For example, programmed death-1 ligand (PD-L)1 and PD-L2 are members of the B7 protein family and are known to bind with programmed death-1 (PD-1) on T cells. Signaling through PD-1 inhibits the activation of T cells by reducing their synthesis of IL- 2 and IFN-γ11, as well as by restricting their entry into the cell cycle12. In recent experiments, blockade of PD-L1 and PD-L2 in antigen-loaded DCs using monoclonal antibodies restored cytokine production by and proliferation of CD4+ T cells in vitro13, and led to the control of ovarian carcinomas in mice following transfusion of ex vivo stimulated T cells14. Furthermore, the DC-derived immunoglobulin receptor 2 (DIgR2) was recently identified and characterised as an immunoinhibitory molecule expressed on the surface of DCs. Vaccination of mice with TAApulsed DCs transfected with siRNA specific for DIgR2 resulted in higher levels of tumour-specific CD4+ and CD8+ T cells as well as protective immunity against tumour challenge15. Additionally, the Notch ligands (Delta1, Jagged1, and Jagged2) are abundantly found on the membranes of DCs and negatively regulate T cell activation and differentiation. It has been revealed that these molecules can coordinate the development of naïve Th cells into different antigen-specific CD4+ T cells, including Th1, Th2, or suppressive T regulatory (Treg) cells16-18. Increased production of IFN-γ was observed in allogenic CD4+ T cells incubated with DCs knocked down for Delta1, Jagged1, and Jagged2 using siRNA19, indicating that Notch ligands may deliver immunoinhibitory signals. These studies signify that PD-L, DIgR2, and Notch ligands are potent suppressors of T cell-mediated responses and that downregulation of these proteins using the RNAi technique should generate improved immunity to tumours.
The recruitment of Treg cells to sites of antigen presentation by DCs serves to control the magnitude of the adaptive immune response through attenuation of T cell activation. Iellem et al. have shown that DCs preferentially attract these immunoregulatory cells by secreting the chemokines CCL17/CCL22, which behave as ligands for CCR4/CCR8 receptors on Treg cells20. Thus, siRNA-mediated silencing of CCL17/CCL22 should reduce Treg cell migration to lymphoid organs and thereby create a microenvironment for antigen presentation that facilitates T cell stimulation.
Preferential selection of a Th1 Cell Immune Response
The cytokine expression profile of activated Th cells is often crucial in determining the success of host immune responses against cancers. Th1 cells can associate with DCs to enhance stimulation of CTLs and also secrete a multitude of immunostimulatory cytokines that boost CTL function. A dominant Th1 response has been shown to be critical for the clearance of the vast majority of tumours. As a result, modifying DCs to select for a Th1 response may be an effective approach for cancer immunotherapy. It is well established that the strength of the Th1 response is critically regulated by the balance between IL-10 and IL-12 signals received by naïve CD4+ T cells during antigen presentation. Transfection of antigen-pulsed DCs with siRNA targeting IL-10 was shown to elevate IL-12 production by these cells and to polarize CD4+ T cell differentiation towards a Th1 profile21. Likewise, it has been reported that IL-6 contributes to the development of a prevalent Th2 response22. Therefore, the use of RNAi technology to reduce IL-6 or IL-10 synthesis by DCs should facilitate the activation of tumour-specific CTLs by amplifying the Th1 response.
The silencing of several of these immunoinhibitory molecules (i.e. SOCS1, DIgR2, PD-L, Notch-L, IDO) with RNAi should enhance antigen presentation and T cell activation. Also, the downregulation of IL-10 and IL-6 synthesis by DCs using this technology should lead to a dominant Th1 response and therefore generate effective CTL-mediated immunity to tumours. Furthermore, Bax/Bak, Fas, and the caspases are crucial in the induction of apoptosis, and thus siRNA-mediated suppression of these proteins in DCs should prolong their life, allowing for sustained antigen presentation and antitumour immune responses.
Prolongation of Dendritic Cell Life
It has been demonstrated that DCs are susceptible to cytolytic granules released by CTLs during antigen presentation23, and that the longevity of DCs is largely determined by their intracellular levels of the antiapoptotic protein Bcl-224,25. Thus, immunotherapeutic strategies that prolong DC life would permit sustained presentation of antigen and should yield synergistic advantages when combined with approaches for enhancing T cell activation. Kim et al. have demonstrated that intradermal coimmunisation of mice with DNA encoding the human papillomavirus- associated oncoprotein E7 in conjunction with Bcl-2 and other apoptosis inhibitors (i.e. Bcl-xL, XIAP, dominant negative caspases-8,9, and SPI-6) via gene gun prolongs DC survival, which correlates with enhanced E7-specific T cell-mediated immune responses and regression of E7-expressing tumours26,27. Delivery of DNA via gene gun has been shown to be an efficient way of introducing DNA directly into DCs. More recently, silencing of proapoptotic proteins (such as Bax/Bak) in DCs using the RNAi technique has been investigated as an alternative method in order to circumvent concerns of oncogenecity associated with the administration of antiapoptotic genes28. It was shown that a DNA vaccine encoding E7 along with Bax/Bak siRNA could be efficiently delivered into DCs in vivo by gene gun administration29. Vaccination of E7-expressing tumour challenged mice elicited strong E7-specific CTL responses as well as anticancer effects29. The success of these experiments both in vitro and in vivo warrant further exploration of the therapeutic potential of DNA vaccines containing TAA along with siRNA targeting the initiator and effector caspases as well as the Fas death receptor.
Prospects for the clinical translation of RNAi technology into a form of cancer immunotherapy
Recent advances in the understanding of tumour biology and immunology have opened up many promising opportunities to treat cancer using the immune system. The rapid development of RNAi technology into a powerful tool for gene silencing over the past few years suggests the exciting possibility of improving cancer vaccines by modifying the properties of DCs to boost their capacity to activate tumour-specific T cells. Nonetheless, despite the remarkable potential for RNA interference in cancer immunotherapy, the major obstacle facing the implementation of this technique in the clinic is the difficulty of efficient in vivo delivery into specific cell types.
Concerns have been raised about the effectiveness of direct systemic administration of siRNA into mammals due to the transient nature of gene silencing, as well as the inherent instability of the RNA molecule. It has been revealed that extracellular siRNA degradation occurs around 36-48 hours after introduction and that the potency of gene knockdown is unpredictable due to the variation in siRNA uptake efficiency by different tissues30. In addition, although the effects of siRNA can last for several weeks in terminally differentiated or senescent cells, they typically expire within one week in rapidly dividing cells. To circumvent these limitations, several groups have developed chemically modified siRNAs that have increased stability and tissue specificity, or are protected from degradation. siRNAs can be altered to resist serum RNase activity, linked with fusogenic peptides, encased in lipid complexes or cationic liposomes, or conjugated to antibodies that bind to cell surface receptors for cell-specific delivery3. Conditionally replicating viruses that express siRNA only in certain cells have also been produced for the cell-specific modification of protein synthesis31. Other strategies to selectively silence genes in particular cell types include use of the highly tissue-specific RNA polymerase II promoter (as opposed to the ubiquitous RNA polymerase III)32 or of stabilised nanoparticles that are preferentially uptaken into target cells33. It is likely that significant progress in the delivery of siRNA into specific tissues will be made in the near future. siRNA can be administered into target cells via viral vectors to achieve more potent gene silencing. These vectors have the ability to integrate into the host genome and induce long-term gene knockdown. Numerous studies have explored viral vector delivery using retroviral34-36, lentiviral37,38, adenoviral38-40 and adeno-associated viral vectors41. Nonetheless, viral vectors generally have low tissue specificity and there are concerns of toxicity associated with the vectors themselves. Some researchers have also developed plasmid-based vectors using RNA polymerase promoters (such as U6 and H1) to express siRNA42. Despite these advances, extracellular stability, tissue-specific delivery and sustained gene silencing remain as significant hurdles that need to be overcome before RNAi technology can be successfully administered in the clinic. Other issues that need to be resolved prior to the therapeutic use of RNAi include potential nonspecific effects, off-target interference and cellular resistance to siRNA43.
As insight into the molecular biological events that mediate RNAi continues to build, it should soon be feasible to safely administer siRNA in vivo to efficiently downregulate proteins in target cells. It would then be possible to implement this technology as an immunotherapeutic intervention to complement traditional cancer vaccines. Also, the silencing of proteins in DCs, T cells, and cancer cells that facilitate tumour evasion of immune responses may lead to the development of novel forms of vaccination that generate optimal and widespread T cell-mediated antitumour responses. Due to the swift progress of research in the biology of cancer, molecules and the immune system, it is expected that RNAi technology may soon translate into a potent form of gene silencing in the clinical arena with profound applications to cancer immunotherapy.


Acknowledgements
This review is not intended to be an encyclopedic one and we apologise to any authors not cited. We thank Shaw-Wei Tsen for his critical review of the manuscript. This article was first published by American Pharmaceutical Review in the August/September issue, 2006.
References
- Caplen, N. J. Gene therapy progress and prospects. Downregulating gene expression: the impact of RNA interference. Gene Ther, 11: 1241-1248, 2004.
- Leung, R. K. and Whittaker, P. A. RNA interference: From gene silencing to gene-specific therapeutics. Pharmacol Ther, 107: 222- 239, 2005.
- Shankar, P., Manjunath, N., and Lieberman, J. The prospect of silencing disease using RNA interference. Jama, 293: 1367-1373, 2005.
- Shen, L., Evel-Kabler, K., Strube, R., and Chen, S. Y. Silencing of SOCS1 enhances antigen presentation by dendritic cells and antigen- specific anti-tumor immunity. Nat Biotechnol, 22: 1546-1553, 2004.
- Kubo, M., Hanada, T., and Yoshimura, A. Suppressors of cytokine signaling and immunity. Nat Immunol, 4: 1169-1176, 2003.
- Zhou, H., Zhang, D.,Wang, Y., Dai, M., Zhang, L., Liu,W., Liu, D., Tan, H., and Huang, Z. Induction of CML28-specific cytotoxic T cell responses using co-transfected dendritic cells with CML28 DNA vaccine and SOCS1 small interfering RNA expression vector. Biochem Biophys Res Commun, 2006.
- Yang, R., Yang, X., Zhang, Z., Zhang, Y., Wang, S., Cai, Z., Jia, Y., Ma, Y., Zheng, C., Lu, Y., Roden, R., and Chen, Y. Single-walled carbon nanotubes-mediated in vivo and in vitro delivery of siRNA into antigen-presenting cells. Gene Ther, 2006.
- Munn, D. H., Sharma, M. D., Lee, J. R., Jhaver, K. G., Johnson, T. S., Keskin,D. B., Marshall, B., Chandler, P., Antonia, S. J., Burgess, R., Slingluff, C. L., Jr., and Mellor, A. L. Potential regulatory function of human dendritic cells expressing indoleamine 2,3-dioxygenase. Science, 297: 1867-1870, 2002.
- Lee, J. R., Dalton, R. R., Messina, J. L., Sharma, M.D., Smith,D. M., Burgess, R. E., Mazzella, F., Antonia, S. J., Mellor,A. L., and Munn,D. H. Pattern of recruitment of immunoregulatory antigen-presenting cells in malignant melanoma. Lab Invest, 83: 1457-1466, 2003.
- Munn, D. H., Sharma, M. D., Hou, D., Baban, B., Lee, J. R., Antonia, S. J., Messina, J. L., Chandler, P., Koni, P. A., and Mellor, A. L. Expression of indoleamine 2,3-dioxygenase by plasmacytoid dendritic cells in tumor-draining lymph nodes. J Clin Invest, 114: 280- 290, 2004.
- Carter, L., Fouser, L. A., Jussif, J., Fitz, L., Deng, B., Wood, C. R., Collins, M., Honjo, T., Freeman, G. J., and Carreno, B. M. PD-1:PDL inhibitory pathway affects both CD4[+] and CD8[+] T cells and is overcome by IL-2. Eur J Immunol, 32: 634-643, 2002.
- Latchman, Y.,Wood, C. R., Chernova, T., Chaudhary, D., Borde, M., Chernova, I., Iwai, Y., Long, A. J., Brown, J. A., Nunes, R., Greenfield, E. A., Bourque, K., Boussiotis, V. A., Carter, L. L., Carreno, B. M., Malenkovich, N., Nishimura, H., Okazaki, T., Honjo, T., Sharpe, A. H., and Freeman, G. J. PD-L2 is a second ligand for PD- 1 and inhibits T cell activation. Nat Immunol, 2: 261-268, 2001.
- Brown, J. A., Dorfman, D. M., Ma, F. R., Sullivan, E. L., Munoz, O., Wood, C. R., Greenfield, E. A., and Freeman, G. J. Blockade of programmed death-1 ligands on dendritic cells enhances T cell activation and cytokine production. J Immunol, 170: 1257-1266, 2003.
- Curiel, T. J., Wei, S., Dong, H., Alvarez, X., Cheng, P., Mottram, P., Krzysiek, R., Knutson, K. L., Daniel, B., Zimmermann, M. C., David, O., Burow, M., Gordon, A., Dhurandhar, N., Myers, L., Berggren, R., Hemminki, A., Alvarez, R. D., Emilie, D., Curiel, D. T., Chen, L., and Zou,W. Blockade of B7-H1 improves myeloid dendritic cell-mediated antitumor immunity. Nat Med, 9: 562-567, 2003.
- Shi, L., Luo, K., Xia, D., Chen, T., Chen, G., Jiang, Y., Li, N., and Cao, X. DIgR2, dendritic cell-derived immunoglobulin receptor 2, is one representative of a family of IgSF inhibitory receptors and mediates negative regulation of dendritic cell-initiated antigen-specific T cell responses. Blood, 2006.
- Hoyne, G. F., Dallman, M. J., and Lamb, J. R. T-cell regulation of peripheral tolerance and immunity: the potential role for Notch signalling. Immunology, 100: 281-288, 2000.
- Wong, K. K., Carpenter,M. J.,Young, L. L.,Walker, S. J., McKenzie, G., Rust, A. J., Ward, G., Packwood, L., Wahl, K., Delriviere, L., Hoyne, G., Gibbs, P., Champion, B. R., Lamb, J. R., and Dallman, M. J. Notch ligation by Delta1 inhibits peripheral immune responses to transplantation antigens by a CD8+ cell-dependent mechanism. J Clin Invest, 112: 1741-1750, 2003.
- Amsen, D., Blander, J. M., Lee, G. R., Tanigaki, K., Honjo, T., and Flavell, R. A. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell, 117: 515-526, 2004.
- Stallwood, Y., Briend, E., Ray, K. M., Ward, G. A., Smith, B. J., Nye, E., Champion, B. R., and McKenzie, G. J. Small Interfering RNA-Mediated Knockdown of Notch Ligands in Primary CD4+ T Cells and Dendritic Cells Enhances Cytokine Production. J Immunol, 177: 885-895, 2006.
- Iellem, A., Mariani, M., Lang, R., Recalde, H., Panina-Bordignon, P., Sinigaglia, F., and D’Ambrosio, D. Unique chemotactic response profile and specific expression of chemokine receptors CCR4 and CCR8 by CD4[+]CD25[+] regulatory T cells. J Exp Med, 194: 847- 853, 2001.
- Liu, G., Ng, H., Akasaki,Y.,Yuan, X., Ehtesham, M.,Yin,D., Black, K. L., and Yu, J. S. Small interference RNA modulation of IL-10 in human monocyte-derived dendritic cells enhances the Th1 response. Eur J Immunol, 34: 1680-1687, 2004.
- Diehl, S. and Rincon, M. The two faces of IL-6 on Th1/Th2 differentiation. Mol Immunol, 39: 531-536, 2002.
- Ingulli, E., Mondino, A., Khoruts, A., and Jenkins, M. K. In vivo detection of dendritic cell antigen presentation to CD4[+] T cells. J Exp Med, 185: 2133-2141, 1997.
- Hou,W. S. and Van Parijs, L. A Bcl-2-dependent molecular timer regulates the lifespan and immunogenicity of dendritic cells. Nat Immunol, 5: 583-589, 2004.
- Nopora, A. and Brocker, T. Bcl-2 controls dendritic cell longevity in vivo. J Immunol, 169: 3006-3014, 2002. American Drug Discovery American Drug Discovery
- Kim, T. W., Hung, C. F., Ling, M., Juang, J., He, L., Hardwick, J. M., Kumar, S., and Wu, T. C. Enhancing DNA vaccine potency by coadministration of DNA encoding antiapoptotic proteins. J Clin Invest, 112: 109-117, 2003.
- Kim, T.W., Hung, C. F., Boyd, D. A., He, L., Lin, C. T., Kaiserman, D., Bird, P. I., and Wu, T. C. Enhancement of DNA vaccine potency by coadministration of a tumor antigen gene and DNA encoding serine protease inhibitor-6. Cancer Res, 64: 400-405, 2004.
- Peng, S., Kim, T.W., Lee, J. H., Yang, M., He, L., Hung, C. F., and Wu, T. C. Vaccination with dendritic cells transfected with BAK and BAX siRNA enhances antigen-specific immune responses by prolonging dendritic cell life. Hum Gene Ther, 16: 584-593, 2005.
- Kim, T.W., Lee, J. H., He, L., Boyd, D. A., Hardwick, J. M., Hung, C. F., and Wu, T. C. Modification of professional antigen-presenting cells with small interfering RNA in vivo to enhance cancer vaccine potency. Cancer Res, 65: 309-316, 2005.
- Ryther, R. C., Flynt, A. S., Phillips, J. A., 3rd, and Patton, J. G. siRNA therapeutics: big potential from small RNAs. Gene Ther, 12: 5-11, 2005.
- Carette, J. E., Overmeer, R. M., Schagen, F. H., Alemany, R., Barski, O. A., Gerritsen, W. R., and Van Beusechem, V. W. Conditionally replicating adenoviruses expressing short hairpin RNAs silence the expression of a target gene in cancer cells. Cancer Res, 64: 2663-2667, 2004.
- Song, J., Pang, S., Lu, Y., Yokoyama, K. K., Zheng, J. Y., and Chiu, R. Gene silencing in androgen-responsive prostate cancer cells from the tissue-specific prostate-specific antigen promoter. Cancer Res, 64: 7661-7663, 2004.
- Schiffelers, R. M., Ansari, A., Xu, J., Zhou,Q., Tang,Q., Storm, G., Molema, G., Lu, P. Y., Scaria, P. V., and Woodle, M. C. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res, 32: e149, 2004.
- Brummelkamp, T. R., Bernards, R., and Agami, R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell, 2: 243-247, 2002.
- Chen, J., Wall, N. R., Kocher, K., Duclos, N., Fabbro, D., Neuberg, D., Griffin, J. D., Shi, Y., and Gilliland, D. G. Stable expression of small interfering RNA sensitizes TEL-PDGFbetaR to inhibition with imatinib or rapamycin. J Clin Invest, 113: 1784-1791, 2004.
- Duxbury, M. S., Ito, H., Benoit, E., Zinner,M. J., Ashley, S.W., and Whang, E. E. Retrovirally mediated RNA interference targeting the M2 subunit of ribonucleotide reductase: A novel therapeutic strategy in pancreatic cancer. Surgery, 136: 261-269, 2004.
- Sumimoto, H., Miyagishi, M., Miyoshi, H., Yamagata, S., Shimizu, A., Taira, K., and Kawakami, Y. Inhibition of growth and invasive ability of melanoma by inactivation of mutated BRAF with lentivirus-mediated RNA interference. Oncogene, 23: 6031-6039, 2004.
- Sumimoto, H., Yamagata, S., Shimizu, A., Miyoshi, H., Mizuguchi, H., Hayakawa, T., Miyagishi, M., Taira, K., and Kawakami, Y. Gene therapy for human small-cell lung carcinoma by inactivation of Skp-2 with virally mediated RNA interference. Gene Ther, 12: 95-100, 2005.
- Chen, L. M., Le, H. Y., Qin, R. Y., Kumar, M., Du, Z. Y., Xia, R. J., and Deng, J. Reversal of the phenotype by K-rasval12 silencing mediated by adenovirus-delivered siRNA in human pancreatic cancer cell line Panc-1.World J Gastroenterol, 11: 831-838, 2005.
- Uchida, H., Tanaka, T., Sasaki, K., Kato, K., Dehari, H., Ito, Y., Kobune, M., Miyagishi, M., Taira, K., Tahara, H., and Hamada, H. Adenovirus-mediated transfer of siRNA against survivin induced apoptosis and attenuated tumor cell growth in vitro and in vivo. Mol Ther, 10: 162-171, 2004.
- Xu,D., McCarty,D., Fernandes, A., Fisher, M., Samulski, R. J., and Juliano, R. L. Delivery of MDR1 small interfering RNA by self-complementary recombinant adeno-associated virus vector. Mol Ther, 11: 523-530, 2005.
- Zheng, L., Liu, J., Batalov, S., Zhou, D., Orth, A., Ding, S., and Schultz, P. G. An approach to genomewide screens of expressed small interfering RNAs in mammalian cells. Proc Natl Acad Sci U S A, 101: 135-140, 2004.
- Pai, S. I., Lin, Y. Y., Macaes, B., Meneshian, A., Hung, C. F., and Wu, T. C. Prospects of RNA interference therapy for cancer. Gene Ther, 13: 464-477, 2006.





[…] de genes que afetam negativamente a sobrevivência e a taxa de apresentação de antígenos pelas células dendríticas (CD). Com isso, é esperado que a taxa de apresentação pelas CD aumente, estimulando positivamente o […]