The role of phospho-proteomics in drug discovery and development
Posted: 25 January 2007 | | No comments yet
While scientific discoveries can be turned into financial assets, the scientific process itself has proven difficult to harness to efficiently create marketed products bringing profits. This translation is especially challenging for the pharmaceutical and biotechnology industries owing to the tremendous complexity of biological systems.
While scientific discoveries can be turned into financial assets, the scientific process itself has proven difficult to harness to efficiently create marketed products bringing profits. This translation is especially challenging for the pharmaceutical and biotechnology industries owing to the tremendous complexity of biological systems.
While scientific discoveries can be turned into financial assets, the scientific process itself has proven difficult to harness to efficiently create marketed products bringing profits. This translation is especially challenging for the pharmaceutical and biotechnology industries owing to the tremendous complexity of biological systems.
In addition to the hope that many new cellular components will become available for pharmaceutical intervention as a result of the genomic revolution, came the challenges of deciphering the biological function of all gene products – the proteins and identifying those, the function of which can be altered chemically to treat a disease. Many years of research that involved thousands of life-scientists across the world working in a field of biological sciences commonly known as ‘signal transduction’, generated enormous amounts of information explaining how the cells sense the various stimuli they receive and how this information is transmitted into the cell interior to ultimately alter its behavior1,2. This information is now being used to create new therapies and therapeutic strategies3. Still, this vast knowledge can be even better leveraged throughout the entire drug discovery and development process.
Using cells to discover new drugs
Some of the advantages of using cells (as opposed to using isolated proteins) to discover new chemical compounds with pharmaceutical value are reducing the need to define the molecular target, creating a larger space of potential drug targets and thus the opportunity to discover chemical structures having unexpected and novel mechanisms of action. Strengthening this view are puzzling situations where qualitative differences are observed between the effects of a chemical compound on an isolated enzyme in-vitro when compared to the effect of such compounds in a bioassay using an intact cell. Post-translational modification of proteins by enzymatic phosphorylation is a pervasive mechanism in cell signaling and perhaps the most widely studied biochemical intracellular reaction3. By modulating the activity of numerous structurally and functionally unrelated proteins, phosphorylation gained involvement in all aspects of cellular life. Hundreds of protein kinases exist and many will phosphorylate in vivo as many as 20-30 distinct substrates. This creates a situation where more than 30 per cent of all cellular proteins encoded by our genome become phosphorylated. Many of them serve as substrates for more than one protein kinase and become phosphorylated on multiple residues. Despite the fact that perhaps thousands of new phosphorylation sites have yet to be discovered, hundreds of phosphorylation sites within cellular proteins have already been identified and, in many cases, their regulatory role has been elucidated. Equipped with this knowledge it now becomes possible to use phosphorylation as an assay endpoint and to obtain valuable information that can be used across the various stages of the drug discovery and development processes.
Applications for phospho-proteomics
Early discovery
In the initial stages of drug-discovery, phosphorylation profiling can help with the choice and characterisation of the cells to be used in the screening of the chemical-compound library. Such profiling also can help to determine whether the signal-transduction pathways that are relevant to the disease are biochemically functional in this cell. It is often important to know whether the cell of interest carries a particular receptor on its surface. While receptor analysis is often an obligatory step, it might be a time-consuming and fairly complicated process. Testing whether the soluble factor can trigger the expected phosphorylation event(s) provides a quick indication that the receptor of interest is not only present at the cell-surface, but also that is found in its functional from and linked to its downstream effectors. This is especially helpful in situations where the number of receptor molecules at the cell-surface is below the detection limit of conventional biochemical techniques or when a cell carries a previously, not described molecular form of the receptor. If a cell-based biological assay produces inconclusive or poorly-reproducible results, biochemical profiling of phospho-events known to mediate or to be associated with this biological process can be used to identify the cell-culture conditions under which the cell is more likely to do better in the biological assay. It also provides the researcher with information that a bioactive factor was used in the study.
Screening
Certain phosphorylation events measured with phospho-specific antibodies (to be discussed later) can serve as an assay endpoint used in the high throughput cell-based screening. A typical example is a program aimed at the discovery of signal-transduction targeted therapeutic to treat malignant diseases. When the goal is to identify an inhibitor of a protein kinase cascade, phosphorylation event occurring a few biochemical steps ‘downstream’ of the target can be used as an endpoint for such an assay. Another example is identification of inhibitors or activators of protein translocation from one cellular compartment to another. Phosphorylation events, in many cases, precede the translocation event and could be used as an assay endpoint and circumvent the requirement for expensive cell-imaging capabilities needed for the high-content screening.
Lead characterisation and optimisation
Phospho-profiling by measuring the phosphorylation state of a relatively small number of key cellular proteins, the regulatory function of which is well established, can be used for assessing compound selectivity and gain an idea of potential toxicities. At this stage, cell-based assays with full dose range of the compound(s) of interest can be performed to create dose-response curves and to obtain in-cell IC50 or EC50 values. This is especially valuable if the primary screening was performed with an in-vitro assay using an isolated target. Such cell-based phosphorylation profiling during an early drug-discovery process may help to distinguish between chemical compounds of similar activity in other types of assays and help in compound prioritisation. It is also possible to use phosphorylation events that compound triggers or blocks to test the relative potency of the compound during the process of the development of its formulation. In cases of protein therapeutic that undergoes modification or reformulation to increase its adsorption, tissue distribution, or serum half-life, this cell-based approach can be used to measure the bioactivity of the modified protein compared to the original unmodified version.
Pre-clinical development/in-vivo studies
Another potential important use for phospho-profiling is biomarker applications in the course of pre-clinical in-vivo studies, such as delineating the PK/PD relationship4. For example, the phosphorylation of signaling molecules such as STATs, SMADs and ERK 1/2 MAP kinase precedes their translocation into the nucleus to initiate transcription events. By looking for phosphorylated forms of these molecules in tissue sections, one may obtain valuable information on the status of the signaling networks represented by these molecules in vivo in control versus treated animals. More recently, phospho-specific antibodies broadly immunoreactive against phosphorylated motifs were developed5 and are now commercially available. Such antibodies, in conjunction with biochemical and proteomic techniques, could be used to discover new assay endpoints to be used in cell-based assays and in in-vivo studies.
Clinical biomarkers
Phospho-specific antibodies can be used for molecular profiling of tumour-tissue biopsies or patient blood samples for diagnostic or prognostic purposes. For example, the ratio between phosphorylated (activated) ERK 1/2 and phosphorylated p38 MAP kinases found to be determinant of tumour growth or dormancy6, while phosphorylation of the protein kinase Akt on Ser473 was found to be a predictor of poor clinical outcome in prostate cancer7. Other types of cellular proteins currently used for diagnostic/prognostic profiling are hyper-phosphorylated cell-surface tyrosine-kinase receptors such as HER2/ErbB2 having an established role in breast cancer8. However, the repertoire of diagnostic/prognostic phospho-protein biomarkers is not necessarily limited to molecules involved in signal transduction. The phosphorylation of such molecules is often transient, resulting in no sufficient accumulation to be detectable in tissue sections. For example, it was found that cytoskeletal and stress proteins are the most abundant tyrosine phospho-proteins in breast tumours9. More recently, phospho-profiling of potentiated signaling networks from cells isolated from patients’ blood was published10. In this case, the isolated blood cells are first stimulated with a panel of cytokines relevant to the disease and then subjected to phospho-profiling. By exposing cancer-cell signaling networks to potentiated inputs, rather than relying upon the basal levels of protein phosphorylation, the researchers could tell the difference between unique profiles that correlated with genetics and disease outcome10.
Discovery of new phosphorylation sites
Overview of methodologies and approaches used to identify new phosphorylation sites
The path leading to the identification of phosphorylation sites utilising classical biochemical techniques used to be a complex, multi-step and time-consuming process. However, even nowadays in some cases, years can pass by before the exact biological role of a particular phosphorylation event and the kinase(s) responsible for it are uncovered. It is estimated that thousands of phosphorylation sites have yet to be discovered and characterised. This article will provide only a bried overview of the three main approaches used for the identification of novel phosphorylation sites.
Traditional biochemistry
The protein is first radiolabeled by enzymatic phosphorylation in-vitro in the presence of radiolabeled ATP and the purified protein kinase suspected to perform the task in-vivo. Alternatively, the protein can be radiolabaled metabolically by adding a radioactive phosphate to the cell growth medium and the cell can then be stimulated with an appropriate stimulus (if necessary) to trigger the phosphorylation event. In the case of metabolic labeling, the phosphorylated protein of interest is first purified from cell extracts either by affinity chromatography or immunotechniques prior to its biochemical manipulation. Then, the radio-labeled phospho-protein is chopped to smaller fragments using proteolytic enzymes having strict and defined specificity. The radioactive protein fragments harbouring the phosphorylated amino acid residue(s) are purified using a variety of chromatographic techniques and their N-terminal amino acid sequence is determined by Edman degradation to reveal their identity. If the radio-labeled fragments are small enough and contain only one amino acid that can possibly become phosphorylated, the identification is fairly clear-cut. Comparison between patterns of radio-labeled protein-fragments obtained from in-vitro phosphorylated protein to the patterns obtained from metabolically labeled cells (peptide mapping) helps to spot the phosphorylation events that are likely to occur in intact cells. Phospho-amino acid analysis is used to reveal whether the phosphorylation occurs on serine, threonine, or tyrosine residues. All in all, this multistep process, parts of which are still in-use by many laboratories, is inefficient, labour-intensive and in many cases produces results that are not easy to interpret.
Mutagenesis
With the advent of the molecular biology era the identification of phosphorylation sites in proteins has been expedited. For this purpose the complimentary DNA corresponding to an open reading frame that encodes the protein of interest must be cloned first into a vector/plasmid. Then site-directed mutagenesis resulting in the substitution of the amino acid suspected to undergo phosphorylation with a non-phosphorylatable amino acid (alanine to substitute serine or threonine and phenyl-alanine to substitute tyrosine) is performed. In order to reduce the number of residues that have to be mutated, a computer-aided search for putative phosphorylation sites in the protein of interest is usually performed first. The knowledge of the preferred amino acid sequence surrounding the phosphorylated residues for many kinases is known and in most cases based on research utilising in-vitro phosphorylation of synthetic peptides, the sequence of which is systematically altered. Optimal or preferred sequences for protein kinases (consensus sequences) are also deduced by comparing the amino acid sequences surrounding multiple previously identified (and published) phosphorylation sites within other proteins. It is worth keeping in mind that the optimal or preferred sequence surrounding the phosporylated residue for many protein kinases is still not known. Putative phosphorylation sites found within domains conserved across species or/and regulatory regions are more likely to be phosphorylated in-vivo and to carry physiological significance.
The approach of introducing point mutations in the putative phosphorylation sites is currently the most popular approach used to uncover new phosphorylation sites and their potential biological role. Its major advantage is that it gives the ability to substitute the amino acid undergoing phosphorylation with an acidic amino acid (aspartic and glutamic for serine or threonine) to mimic the addition of the negatively charged phosphate group. This means that functional cell-based assays can be performed to investigate the effect of the substitution on the biological process of interest. Some problems are encountered in cases where several tandem phosphorylation sites are present at a close proximity to each other and become phosphorylated in an ordered and hierarchical manner by one or more kinases. The introduced point mutations may alter the sequence in the context of which the protein kinase recognises the phosphorylation site in the mutated protein. A typical example is ‘priming phosphorylation’ where the kinase requires a neighboring pre-phosphorylated site. Perhaps the major drawback of the molecular approach is that it requires an ectopic expression by transfecting the cell with a plasmid encoding the mutated protein of interest. Ectopic expression may lead to accumulation of the protein within a sub-cellular compartment in which it naturally does not reside leading potentially to artificial phosphorylation events and effect on unrelated cellular functions.
Mass-spectrometry
Identification of phosphorylation sites by mass spectrometry relies on the proteolytic cleavage of proteins (either phosphorylated in-vitro or purified from stimulated cells) into short peptides, ionisation of the peptides either by electrospray (ESI) or matrix-assisted laser desorption (MALDI) and accurate peptide mass determination11. For single proteins, a comprehensive peptide map obtained by MALDI-TOF (TOF- Time of flight) is often sufficient to unambiguously identify the protein by a database search of all possible tryptic peptides. For more complex samples tandem mass spectrometry (MS/MS) is used11. This method introduces the additional step of peptide ion fragmentation at the amide bonds, which directly yields protein sequence information. The presence of a phosphate group on a peptide is determined by a predicted increase in peptide mass due to addition of the phosphate group. A farther improvement to the technique was achieved by enriching the samples for phosphorylated proteins or phospho-protein fragments derived from cell extracts utilising phospho-specific antibodies or metal-affinity chromatography12-15. The systematic detection of protein phosphorylation remains a challenge for current mass spectrometric methods, in part because phosphoserine and phosphothreonine residues are very labile and often lost during the peptide fragmentation step. The main benefit from applying this technology is the ability to discover novel phosphorylation sites occurring in their natural biological context. It does come with a price – mass spectrometry requires expensive specialty equipment, skilled technical personnel, labour-intensive sample handling and produces large data sets.
Antibody based approaches to study the phospho-proteom
Because of their remarkable capability for molecular discrimination, specific biorecognition and versatility antibodies found a broad usage in a large variety of applications ranging from basic biomedical research through clinical diagnostics to therapeutic intervention. Phosphorylation-state specific antibodies proved to be a powerful tool not only for studying signal-transduction but also in investigating protein phosphorylation in situ, thus opening many exciting opportunities in diagnostic pathology16,17. More than two decades ago, phospho-specific antibodies immunoreactive with phosphorylated tyrosines were developed18. Such antibodies are still in use today and will immunoreact with any protein phosphorylated on tyrosine residue(s) regardless of the amino acid sequence that surrounds it. Ten years later, the development of phospho-specific antibodies against proteins phosphorylated on serine or threonine was published19. These antibodies immunoreact with phosphorylated residues only when found in the context of a defined amino acid sequence that surrounds them. The major limitation of applications that rely on phospho-specific antibodies is that prior identification of the phosphorylation site, as well as the generation and validation of the phospho-specific antibody, are required.
Nowadays, the generation of phosphorylation-state specific antibodies is routinely achieved through chemical synthesis of a short peptide having a sequence corresponding to the amino acid sequence surrounding the phosphorylation site within a cellular phospho-protein of interest. This peptide is synthesised with the appropriate serine, threonine, or tyrosine side-chains to which a phosphate group is covalently attached through chemical synthesis to mimic the naturally occurring phosphorylation. After its purification, the phospho-peptide is chemically conjugated to a carrier protein and is used to immunise rabbits, mice, or goats utilising standard immunisation protocols. The phospho-specific antibody is affinity purified by sequential chromatographic steps using first non-phospho peptide then phospho-peptide affinity columns. In many cases, the resultant antibodies will immunoreact with the target protein only when the particular serine, threonine, or tyrosine is phosphorylated.
Often, cellular proteins become phosphorylated on multiple sites (serine, threonine, tyrosine) by a variety of protein kinases, with each phosphorylation having potentially a unique consequence for this protein’s function. Phospho-specific antibodies make it possible to monitor the phosphorylation state of defined residues within a single protein. Because of increasing use of phospho-specific antibodies in basic biomedical research both by academic and industrial researchers, many manufacturers of research-enabling products offer now a large panel (several hundreds are now commercially available) of such phospho-specific antibodies. To fully benefit from an analysis using phospho-specific antibodies, the biological function of the measured phosphorylation event must be known.
Some significant advantages of using phospho-specific antibodies are that it does not necessarily require expensive specialty equipment and valuable data can be obtained using standard biochemical procedures (such as western-blots) that are not encumbered by patent restrictions. In many cases, the phosphorylation state of an endogenous cellular protein can be measured meaning that ectopic expression of the protein (by introducing a foreign gene using recombinant DNA technology) can be avoided. Therefore, the approach allows monitoring important molecular events that occur in their natural cellular context and are likely to reflect true biological processes.
Like other biochemical and cell based-assay formats, antibody based assays have the capability to interface with a number of ubiquitous drug discovery technological platforms. These usually involve any combination of the following: surface chemistry for immobilisation, protein array spotters, liquid handling using multi-channel robotic dispensers/washers, plate readers/ recorders, cell-imaging/microscopy and data acquisition/data analysis software. Proper combination and usage of these technologies allow the conversion of an antibody based assay to a high throughput mode and/or enable the simultaneous measurement of several reactions – multiplexing (reducing cost and time). One prominent way of increasing the throughput is surface immobilisation of antibodies using either direct spotting or surface chemistries that allow oriented immobilisation. The immobilisation can be on a variety of supports ranging from nitrocellulose membranes, glass slides or the bottom of multiwell micro plates. The latter especially is advantageous since it allows the conversion of an assay to a high throughput format by using existing robotic liquid handling instruments interfacing with standard plate readers having fluorescence and/or luminescence capabilities. In such an assay format akin of sandwich ELISA, the surface immobilised antibody is used to capture the protein of interest from the complex sample of cell extract. After binding and washing the non-specifically bound material a second antibody (fluorescently labeled or enzyme-linked) immunoreacting with a distinct region on the captured protein is used to quantify the bound material. This means each protein of interest is positively identified by a pair of antibodies in which one of them is phospho-specific.
Another prominent technology utilising antibodies that has recently gained popularity is immobilisation on a suspension array of colour-coded beads that are sorted according to a principle very similar to the classical flow-cytometry technology – FACS (Fluorescence Assisted Cell Sorting). Antibodies with different specificities are immobilised on beads, each having a unique colour code (visible to an instrument, not the human eye). The colour-coded bead suspension with immobilised antibodies (phospho-specific) is mixed, the analyte (i.e., cell extract) is added and the proteins of interest are captured on beads carrying antibodies. Next, a second set of fluorescently labeled antibodies reacting with the phospho-proteins on a surface distal from the first set antibodies (up to a dozen in a single reaction) is added resulting in each phospho-protein of interest being tagged by a pair of non-redundant antibodies. The liquid sample with bead suspension is fed into the instrument which can determine the relative number of beads of each colour and the relative intensity of fluorescence attached to them originating from the second antibody. This intensity is directly proportional to the amount of the phospho-protein in question. The main advantage of this technology is the lack of need for washing to eliminate the non-specific binding. This assay format is functionally homogeneous but requires considerable assay optimisation process to achieve optimal results and proper robustness.
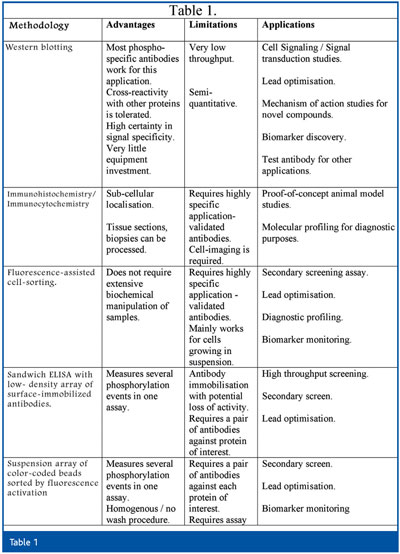
Recently, a measurement of phospho-proteins in cells was described utilising the classical flow-cytometry methodology using fluorescently labeled, phospho-specific antibodies20,21. The weak spot of this approach is that the protein of interest within an intact cell has to be identified by a single antibody as opposed to an antibody pair in the previous two technologies mentioned above. This hurdle can be overcome by using highly selective antibodies that were validated for this application. Another interesting possibility for increasing the throughput that has yet to be experimentally demonstrated is to use the phospho-specific antibodies in Immunoaffinity Capillary Electrophoresis (IACE)22. This technique combines the powers of the antibody antigen specific biorecognotion with that of the electrophoretic separation and has the potential for high throughput and automation. Comparison between the various technologies that rely upon phospho-specific antibodies and their potential uses are described in Table 1.

References
- Thomas, G.d.P., F; Schlessinger, J; Moscat, J, The ins and outs of protein phosphorylation. Workshop report: control of signaling by protein phosphorylation. EMBO Reports. 2000 Jul;1(1):11-5., 2000. 1(1): p. 11-15.
- Downward, J., The ins and outs of signalling. Nature, 2001. 411(6839): p. 759-762.
- Cohen, P., The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. European Journal of Biochemistry, 2001. 268(19): p. 5001-510.
- Espina, V.D., KA; Cowherd, S; Petricoin, EF 3rd; Liotta, LA, Use of proteomic analysis to monitor responses to biological therapies. Expert Opinion in Biology and Therapy, 2004. 4(1): p. 83-93.
- Zhang, H.Z., X; Tan, Y; Hornbeck, PV; Mastrangelo, AJ; Alessi, DR; Polakiewicz, RD; Comb, MJ., Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. Journal of Biological Chemistry, 2002. 277(42): p. 39379-39387.
- Aguirre-Ghiso, J.E., Y; Liu, D; Ossowski, L, ERK(MAPK) activity as a determinant of tumor growth and dormancy; regulation by p38(SAPK). Cancer Research, 2003. 63(7): p. 1684-1695.
- Kreisberg, J.M., SN; Prihoda, TJ; Bedolla, RG; Troyer, DA; Kreisberg, S; Ghosh, PM, Phosphorylation of Akt (Ser473) is an excellent predictor of poor clinical outcome in prostate cancer. Cancer Research, 2004. 64(15): p. 5232-5236.
- Ouyang, X.G., T; Doherty, A; Huang, GC; Epstein, RJ, Detection of ErbB2 oversignalling in a majority of breast cancers with phosphorylation-state-specific antibodies. Lancet, 1999. 353(9164): p. 1591-1592.
- Lim, Y.W., CY; Ooi, LL; Druker, BJ; Epstein, RJ, Selective tyrosine hyperphosphorylation of cytoskeletal and stress proteins in primary human breast cancers: implications for adjuvant use of kinase-inhibitory drugs. Clinical Cancer Research, 2004. 10(12 (part 1)): p. 3980-3987.
- Irish, J.H., R; Krutzik, PO; Perez, OD; Bruserud, O; Gjertsen, BT; Nolan, GP, Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell, 2004. 118(2): p. 217-228.
- Ashman, K.M., MF; Sicheri, F; Pawson, T; Tyers M, Cell Signalling – The Proteomics of It All. Science’s STKE, 2001. http://stke.sciencemag.org/cgi/content/full/OC_sigtrans;2001/103/pe33.
- Gronborg, M.K., TZ; Stensballe, A; Andersen, JS; Ohara, O; Mann, M; Jensen, ON; Pandey, A, A mass spectrometry-based proteomic approach for identification of serine/threonine-phosphorylated proteins by enrichment with phospho-specific antibodies: identification of a novel protein, Frigg, as a protein kinase A substrate. Molecular and Cellular Proteomics, 2002. 1(7): p. 517-527.
- Ficarro, S.M., ML; Stukenberg, PT; Burke, DJ; Ross, MM; Shabanowitz, J; Hunt, DF; White, FM, Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nature Biotechnology, 2002. 20(3): p. 301-305.
- Salomon, A.F., SB; Brill, LM; Brinker, A; Phung, QT; Ericson, C; Sauer, K; Brock, A; Horn, DM; Schultz, PG; Peters, EC, Profiling of tyrosine phosphorylation pathways in human cells using mass spectrometry. Proceedings of the National Academy of Science U S A., 2003. 100(2): p. 443-448.
- Brittain, S.F., SB; Brock, A; Peters, EC, Enrichment and analysis of peptide subsets using fluorous affinity tags and mass spectrometry. Nature Biotechnology, 2005. 23(4): p. 463-468.
- Nagata, K.I., I; Inagaki, M, A decade of site- and phosphorylation state-specific antibodies: recent advances in studies of spatiotemporal protein phosphorylation. Genes and Cells, 2001. 6(8): p. 653-664.
- Mandell, J.W., Phosphorylation state-specific antibodies: applications in investigative and diagnostic pathology. American Journal of Pathology, 2003. 165(5): p. 1687-1698.
- Ross, A.B., D; Eisen, HN, Phosphotyrosine-containing proteins isolated by affinity chromatography with antibodies to a synthetic hapten. Nature, 1981. 294(5842): p. 654-656.
- Czernik, A.G., JA; Nairn, AC; Chen, J; Snyder, G; Kebabian, J; Greengard, P, Production of phosphorylation state-specific antibodies. Methods in Enzymology, 1991. 201: p. 264-283.
- Chow, S.P., H; Hedley, DW, Measurement of MAP kinase activation by flow cytometry using phospho-specific antibodies to MEK and ERK: potential for pharmacodynamic monitoring of signal transduction inhibitors. Cytometry, 2001. 46(2): p. 72-78.
- Krutzik, P.N., GP, Intracellular phospho-protein staining techniques for flow cytometry: monitoring single cell signaling events. Cytometry Part A, 2003. 55(2): p. 61-70.
- Guzman, N.P., TM, Immunoaffinity Capillary Electrophoresis for proteomics studies. Analytical Chemistry 77(3):61A-67A, 2005. 77(3): p. 61A-67A.
Acknowledgement
This article was originally published in American Drug Discovery, Volume 1, Issue 1, 2006.




