Polyphasic approach to microbial identification
Posted: 28 November 2006 | | No comments yet
The identification of microorganisms from the pharmaceutical production environment has gained an ever greater importance in modern times. Thus the new Aseptic Processing Guide of the FDA recommends the identification of detected isolates from the critical clean room area (grade A or ISO 5) down to the species level and recommends the identification of isolates from the surrounding lesser controlled clean room area (i.e. grade B or ISO 7) down to at least the genus level.
The identification of microorganisms from the pharmaceutical production environment has gained an ever greater importance in modern times. Thus the new Aseptic Processing Guide of the FDA recommends the identification of detected isolates from the critical clean room area (grade A or ISO 5) down to the species level and recommends the identification of isolates from the surrounding lesser controlled clean room area (i.e. grade B or ISO 7) down to at least the genus level.
The identification of microorganisms from the pharmaceutical production environment has gained an ever greater importance in modern times. Thus the new Aseptic Processing Guide of the FDA recommends the identification of detected isolates from the critical clean room area (grade A or ISO 5) down to the species level and recommends the identification of isolates from the surrounding lesser controlled clean room area (i.e. grade B or ISO 7) down to at least the genus level.
For this purpose the microbiological quality control laboratory can use a multitude of methods. This article discusses exemplarily the established procedures that are used by the Novartis Pharma Stein AG for the identification of microbial isolates from the pharmaceutical production environment.
The identification of microorganisms poses an ever increasing challenge for the microbiological quality control laboratory, because the demand for precise identifications have steadily increased and because the personnel handling the identification need both good microbiological training and detailed expertise to be able to evaluate the plausibility of the results of the mostly automated identification systems.
From the viewpoint of authorities the increasing demands for identification are understandable, because the identification of microorganisms provides an important clue for the identification of a potential contamination source and can help explain deviating results of microbiological analyses. For example, confirming detection of Staphylococcus epidermidis in a sample of water for injection (WFI) that is used for the manufacture of a drug product is a good indicator of a potential contamination source during sampling or the processing of the sample in the laboratory. Furthermore, microbial identification together with the comparison of results enables detection of negative trends; i.e. the occasional contaminations with a microorganism found in environmental controls of a critical clean room area (i.e. grade A or ISO 5) can indicate the introduction of this microorganism from surrounding lesser controlled production areas.
Moreover, the number of identifications that must be processed has steadily increased and this, together with the refined methods for identification, raises questions of the cost-efficiency of the process. Therefore, the quality control laboratory has to make a decision whether or not every isolate should be identified to the species level2.
In addition, the ever shortening cycle-times of the analysis influences the microbial identification, especially in connection with the PAT initiative3, that leads to a faster release of the manufactured products; thus automated systems usually result in a higher throughput of identifications.
To meet all these requirements the team at Novartis Pharma Stein AG, a production location for sterile and solid pharmaceutical forms, have set up the following procedure for microbial identification (see below).
Number of identifications
To minimise the number of identifications it is necessary to define a concept for microbial identification. Generally, all colonies of a plate are identified that can be morphologically unambiguously differentiated. Thereby all morphological forms from the critical clean room areas grade A and B (ISO 5 and ISO 7) of the microbial environmental controls will be identified, independently of whether the warning or action limit is exceeded. In all other microbial examinations an identification will only be made after exceeding the warning limit (i.e. examination of pharmaceutical water) or the action limit (i.e. clean room areas grade C and D). Depending on the status of hygiene in the clean room areas, the described procedure enables a drastic reduction in the number of microbial identifications, since the microbial environmental controls are the largest share of the overall microbial identifications.
Isolation of microorganisms
The identification should generally be carried out beginning with a pure culture. This means that as a first step for identification a sub-cultivation on a suitable medium must be made. It is therefore necessary to plate a single colony on a medium that is required for further identification. This procedure has the possibility to be repeated several times in order to obtain a pure culture (uniform colony type with respect to colour, form, etc.). Although molecular biological methods (PCR-based amplification) generally allow the identification from the original test plate without sub-cultivation, they run the risk of qualitative poor identification results (high percentage of mixed bases in the sequence) because potentially overlapping growing colonies were not separated.
Pre-differentiation
The pre-differentiation usually consists of a Gram-stain followed by the microscopic analysis of the stained specimen, complemented by biochemical tests or further staining in case the Gram-stain does not show an unequivocal result. The continuing tests are catalase, oxidase, KOH-test, amino-peptidase, spore staining and oxidation-/fermentation-(OF-) test. The microscopic analysis of the Gram-staining results in additional information about the morphology of the examined isolates (rod-shaped or coccoid bacteria, spores, vibrioid, or form-variable cells, yeasts).
The pre-differentiation is an extremely critical step in the whole identification process, especially using phenotypic identification systems4,5. These identification methods show a certain variance of the results depending on the duration of the sub-cultivation, the used medium and the incubation temperature. Therefore, it is essential for the quality of the identification results of these methods to use as standardised sub-cultivation conditions as possible with regards to the temperature and the duration of the sub-cultivation. In addition, the number of media is limited, which in certain cases terminates the identification already at the stage of the sub-cultivation for the pre-differentiation, since insufficient microbial growth occurs. According to experience, this is often the case with microorganisms from pharmaceutical water. These microorganisms are adapted to minimal nutritional conditions so that the microorganisms can potentially only accomplish the change to a full medium (i.e. TSA Tryptic Soy Agar) after a prolonged incubation phase. Cultures of these bacteria in ultra-pure water without additional carbon source show (even over a prolonged time) a stable cell count and do not die6; a phenomenon that documents their elaborate adaptation to this difficult habitat.
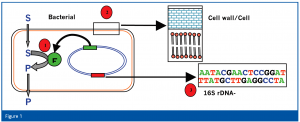
The results of the pre-differentiation determine the selection of the corresponding microbial identification systems. We therefore use three different identification systems; two of which are based on the determination of phenotypic characteristics, while the third is based on the analysis of genotypic characteristics (Figure 1).
Bacteria from the group of the Enterobacteria (Gram-negative, oxidase negative, OF-test positive) are further analysed with a biochemical-based identification system.
All other bacteria are analysed using the gas chromatographic analysis of the fatty acid profile of the bacterial cell membrane. Depending on the pre- differentiation, various methods are used for the sub-cultivation of the microbe.
All yeasts, fungi and bacteria that cannot be identified using the first two methods are identified using a molecular biology-based identification system. Additionally, the identification of isolates from product release tests or validation runs of the filling machine (i.e. sterility tests, microbial limit test, media fill) is generally achieved using the molecular biology identification method.
Biochemical identification methods
Various instruments of different manufacturers exist for identification using biochemical characteristics (i.e. metabolism of different carbon sources). The best-known ones are the API® and VITEK® Microbial Identification Systems (bioMérieux S.A., Marcy-l’Etoile, France) and the Omnilog® Microbial Identification System (Biolog, Hayward, California USA). For the identification of Enterobacteria in our laboratory, the API System (ID 32 E) is used in combination with the online database API Web. The API System offers advantages for the identification of Enterobacteria with respect to the discrimination of individual species, whereas the identification of Enterobacteria with a genotypic system is very difficult, because the 16S rRNA sequences of individual species can differ in only a few bases. A good example is Escherichia coli ATCC 11775 and Shigella dysenteriae ATCC 13313 that differ genotypic in only three bases of their 16S rRNA gene, so that they could potentially be viewed as one species, even though clinically and historically they are clearly two different species7.
The disadvantages of the biochemical identification methods are a long incubation period, the dependency of the growth of the test culture from the quality of the pre-culture (cells during the logarithmic growth phase are empirically the best inoculum) and the limitation that many organisms cannot grow on the used media. In addition, many of the bacterial entries in the biochemical identification systems are based on clinical bacteria, not environmental bacteria.
Identification by gas chromatographic analysis of Fatty Acid Methyl Esters (FAME)
For routine bacterial identification, we use the MIDI Sherlock® Microbial Identification System (MIDI, Inc, Newark, Delaware USA) in our laboratory. This system differentiates bacterial species based on the unique fatty acid composition of each bacterial cell membrane, after growth on standardised media using defined conditions. Fatty acids are isolated from the cells, saponified, methylated (to form fatty acid methyl esters) and then separated using gas chromatography. The species-specific fatty acid profile is subsequently compared with a database and evaluated for the closest match(es). The pattern recognition software calculates a similarity index (SI) that statistically quantifies the match between the unknown fatty acid profile and the fatty acid profiles in the Sherlock databases to determine the closest match(es) (Figure 2).
Sample processing for the gas chromatograph is fully automated and allows for a high sample throughput with a low material effort and therefore at a low cost basis (agar plates for the sub-cultivation, a few common laboratory chemicals for the sample preparation). Furthermore, the implementation of the new Sherlock version 6.0 Rapid Aerobe Method/Library has helped to establish a rapid method capable of identifying a large number of environmental bacterial species. The time necessary to perform identifications using the gas chromatograph was reduced to nine minutes from 27 minutes with the Standard Aerobe Method.
The disadvantage of the method is the long incubation time for the sub-culture and the limited number of growth media suited for the sub-cultivation of the bacteria. This results in the difficulty that certain bacteria cannot be cultivated on the specified media because of their nutritional requirements and, therefore, cannot be identified. For example, sub-cultivation of bacteria that have been isolated on R2A on the recommended medium TSBA results in very long incubation times. Similarly, the growth of Corynebacterium species on blood agar is very slow, which often results in an insufficient amount of cell material for optimal analysis of the fatty acid composition.
Furthermore, the pre-differentiation is of importance for the selection of the medium (TSBA or blood agar) and the selection of the used database for analysis (CLIN-clinical or TSBA-environmental database). Therefore, the pre-differentiation must be carried out by experienced and well-trained laboratory personnel. An erroneous pre-differentiation can result in an identification that does not correlate with the result of the pre-differentiation and instigates further investigation.
Despite these limitations, however, fatty acid analysis by gas chromatographic separation is very easy to perform, reproducible and inexpensive. Moreover, in a majority of cases (isolates from microbial environmental controls) fatty acid analysis leads to satisfactory results owing to the very large environmental databases. At least the identification down to the genus level (and in many cases species level) is possible.
MIDI has recently introduced two new software options geared to pharmaceutical industrial applications; Sherlock® Tracker (capable of real-time strain tracking) and Sherlock® DNA (which allows for the combination of 16S rRNA sequencing and fatty acid data into a combined polyphasic result), but the microbiological quality control laboratory has not evaluated either of these software options yet.
Identification using 16S rRNA gene sequencing
The identification portfolio is ideally complemented through the establishment of a genotypic identification method, which allows a further identification possibility for isolates that cannot be unambiguously identified using phenotypic methods. The method used is the MicroSeq® Microbial Identification System (Applied Biosystems, Foster City, California USA) and has only been available for a few years with the support of comprehensive databases from Applied Biosystems (500bp-, full gene- and fungal-database) for pharmaceutical industrial applications. In addition to the identification of bacteria, the MicroSeq® System offers the identification of yeasts and molds using a database that is more extensive than that of other identification systems. This possibility offers, for many quality control laboratories, an alternative to the phenotypic or purely morphologic identification of molds that in the case of a morphologic identification can only be reasonably accomplished with experience and the application of complex identification keys. Despite the fact that the yeast and mold database still requires expansion7,10,11, it already offers more options for a reliable identification of yeasts and molds than a purely morphological approach.
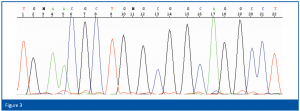
For identification, the first 500 bp of the bacterial 16S rRNA gene or the 300 bp long D2 region of the rRNA gene of the large ribosomal subunit of yeasts and molds are amplified by PCR using universal primers and the resulting fragments are subsequently sequenced. For the isolation of the chromosomal DNA, the PCR reaction and the cycle-sequencing reaction, corresponding kits with enzymes, buffers, primers etc. are available. No further reagents are necessary. The determination of the sequence is automatically carried out using a capillary electrophoresis unit (an example is shown in Figure 3). The resulting sequence is then compared with the database sequences and the results expressed in per cent match.
The difficulty with the identification process results from the determination of a cut-off for the identification, i.e. how many percentage sequence identities are necessary to identify the species or the genus level, respectively. The average sequence difference of the 16S rRNA-gene of different species of a bacterial genus can vary widely. For example, the average sequence difference between species of the genus Pseudomonas is 9.4%, while the differences within the genus Enterobacter are 2.1% and within the genus Aeromonas, 1.0% (8 supplemented from 7). These different values make it difficult to define a general cut-off value for reliable species identifications. Based on the performed pre-differentiation we have chosen a cut-off value of >98% sequence identity for our species identifications and have so far had very good experiences (no non-plausible results between pre-differentiation and MicroSeq®-based results). A more stringent value for sequence homology is recommended, should the method be used without preceding pre-differentiation.
The advantage of this method is the independence from the cultivation conditions; meaning that the genetic information is unchanged and independent of the environment in which the microorganism grows. This is confirmed by a study that showed that the PCR-profile of different bacteria was stable over several cycles of sub-cultivation9. Similarly, we could show in our laboratory during the initial validation of the system that identical results were obtained independently, whether the identification started from an original plate or from a sub-culture.
The disadvantage of this method is the relatively expensive upfront cost and high price per test.
Summary and outlook
Using the procedures and technical equipment described, the microbiological quality control laboratory is able to plausibly obtain (for a large number of identifications (>98%)) the species, or at least the genus level identification, irrespective of which microbiological analysis the corresponding isolate came. The few cases in which the identification has to be concluded with the result ‘not-identifiable’ are the consequences of gaps within the used databases. This observation is not too astonishing, given the extreme large number of bacteria that, until now, have been taxonomically classified using genetic data, but have not yet been cultivated or are un-cultivable12. Estimates about microbiological diversity differ widely. The fact is that, until now, only a small percentage of existing species have been described in detail13. Therefore, it is not surprising that even in the pharmaceutical production environment, new species are found on a continual basis that cannot (or only unsatisfactorily) be identified using existing methods. For these reasons, our current microbial systematic will further adjust in the future, what, on the other side, will lead to a continuous development and expansion of our identification systems and their databases.




References
- Guidance for Industry: Sterile drug products produced by aseptic processing – Current good manufacturing practice; U.S. Food and Drug Administration: Rockville, MD, 2004.
- Seyfahrt H. Microbiological monitoring, Part 6: Identification of isolates from environmental controls. Pharm. Ind. 2004; 66 (11):1358 – 1363.
- Guidance for Industry: PAT – A framework for innovative pharmaceutical development, manufacturing, and quality assurance; U.S. Food and Drug Administration: Rockville, MD, 2004.
- Sutton SVW, Cundell, AM. Microbial identification in the pharmaceutical industry. Pharmacopeial Forum 2004; 30 (5): 1884 – 1894.
- Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral JP, Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 2000; 38 (10): 3623 – 3630.
- McAlister MB, Kulakov LA, O’Hanlon JF, Larkin MJ, Ogden KL. Survival and nutritional requirements of three bacteria isolated from ultrapure water. J. Ind. Microbiol. Biotechnol. 2002; 29: 75 – 82.
- Clarridge III JE. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 2004; 17 (4): 840 – 862.
- Montgomery S, Anderson S, Waddington M, Bartell J, Nunn G, Foxall P. Variation in bacterial interspecific genetic distances – New rules for interpretation of 16S rDNA sequences? In Program and abstracts of the IXth International Congress of Bacteriological and Applied Microbiology, Sydney, Australia, 1999.
- Kang HP, Dunne WM. Stability of repetitive-sequence PCR patterns with respect to culture age and subculture frequency. J. Clin. Microbiol. 2003; 41 (6): 2694 – 2696.
- Hall L, Wohlfiel S, Roberts GD. Experience with the MicroSeq D2 Large-Subunit ribosomal DNA sequencing kit for identification of filamentous fungi encountered in the clinical laboratory. J. Clin. Microbiol. 2004; 42 (2): 622 – 626.
- Hall L, Wohlfiel S, Roberts GD. Experience with the MicroSeq D2 Large-Subunit ribosomal DNA sequencing kit for identification of commonly encountered, clinical important yeast species. J. Clin. Microbiol. 2003; 41 (11): 5099 – 5102.
- Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu. Rev. Microbiol. 2003; 57: 369 – 394.
- Ammann R. Who is out there? Microbial aspects of biodiversity. System. Appl. Microbiol. 2000; 23: 1 – 8.




