A showcase project for EU research
Posted: 20 July 2006 | | No comments yet
The investigation of functional protein-protein interactions has been gaining momentum with recent technological innovations. The high-throughput era in genomics and proteomics research is essentially dependent on technological advancements to drastically increase capacities in both large-scale gathering of data; their interpretation and functional validation, as well as the compilation and storage of data in a standardised format. Contributions of the European Commission-funded ‘Interaction Proteome’, an integrated project in the field, are outlined in this article.
The investigation of functional protein-protein interactions has been gaining momentum with recent technological innovations. The high-throughput era in genomics and proteomics research is essentially dependent on technological advancements to drastically increase capacities in both large-scale gathering of data; their interpretation and functional validation, as well as the compilation and storage of data in a standardised format. Contributions of the European Commission-funded ‘Interaction Proteome’, an integrated project in the field, are outlined in this article.
The investigation of functional protein-protein interactions has been gaining momentum with recent technological innovations. The high-throughput era in genomics and proteomics research is essentially dependent on technological advancements to drastically increase capacities in both large-scale gathering of data; their interpretation and functional validation, as well as the compilation and storage of data in a standardised format. Contributions of the European Commission-funded ‘Interaction Proteome’, an integrated project in the field, are outlined in this article.
The Interaction Proteome project was initiated in 2004 with the ambitious goal to improve the state-of-the-art in all the above areas. Now, at mid-term of the project’s lifetime, an assessment of the achievements is due.
2D-Gel-free proteomics technologies
Recent innovations in the field of mass spectrometry render the classical and well established approach, including a separation of proteins via two-dimensional (2D) gel electrophoresis, increasingly dispensable. Novel 2D-Gel-free technologies are superior with respect to a direct connection of protein separation and analysis via mass spectroscopy along with the feasibility of automation.
A major breakthrough for the field was the development of a novel mass spectrometer, the LTQ-Orbitrap and its introduction to the market in June 2005 by Interaction Proteome partner Thermo Electron, Bremen. The Orbitrap is the first fundamentally new mass analyser in more than 20 years. It exhibits a ten-fold increase in sensitivity along with a four-fold extension of the dynamic range as compared to a state-of-the-art high performance mass spectrometer (LTQ-FT). Based on the highly accurate mass determination combined with high resolution and sensitivity, the novel instrument not only allows for the routine analysis with high-throughput, but also for the straight forward analysis of peptide mixtures without chemical or enzymatic modifications1. The Orbitrap thus is the ideal instrument for the two novel 2D-Gel-free proteomics approaches developed within the project, namely the SILAC (Stable Isotopic Labelling by Amino acids in Cell culture) technology by the group of Matthias Mann, University of Odense/Max Planck Institute of Biochemistry, Martinsried and the COFRADICTM (COmbined FRActional DIagonal Chromatography) approach by the group of J. Vandekerckhove, Flanders Interuniversity Institute of Biotechnology, Ghent.
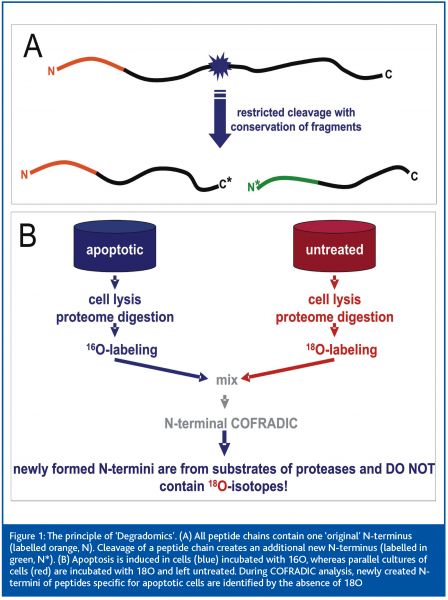
COFRADIC uses chemical and/or enzymatic modification (for example of rare amino acids or the N-terminus) between two identical chromatography steps to select peptides whose running behaviour changes characteristically in subsequent analysis. An extension of this method has been successfully used in an approach on Degradomics – the analysis of protein processing in apoptosis. This morphologically and physiologically defined form of cell death involves specific cleavage of protein sets in the cell. This releases fragments with newly created N-termini, which are detected by the N-terminal COFRADIC technology (Figure 1A). The comparative analysis of peptide lysates of 18O labelled untreated cells with cells labelled with 16O upon induction of apoptosis (Figure 1B) enabled the identification of over 90 protease substrates and the exact localisation of the cleavage sites. This study2 provided the first comprehensive analysis of the “apoptotic degradome” and a proof of principle for the analysis of protein processing in a wide variety of biomedically relevant systems.
In parallel, SILAC technology was extended towards a mass-spectrometry based quantitative proteomics approach to investigate signalling cascades in mammalian cells. It was used to characterise the role of Receptor Tyrosine kinases (RTK) in the differentiation of mesenchymal stem cells to bone cells. The two RTK ligands’ epidermal growth factor (EGF) and platelet-derived growth factor (PDGF) elicit differential effects in the stem cells; whereas PDFG induces motility and survival, EGF treatment leads to differentiation towards mature bone cells. A comparison of RTK signalling complexes induced by EGF and PDGF in mesenchymal stem cells revealed that both growth factors activated a largely overlapping set of signalling proteins, whereas only PDGF induced the phosphatidylinositol 3-kinase (PI3K) pathway. Subsequent in vitro and in vivo experiments confirmed that the activity of the PI3K pathway inhibited the differentiation process. This study3 shows that global quantitative proteomics is a suitable tool to compare closely related signalling networks and to identify critical differences in their signalling capacities. The characterisation of pathways affecting cell fate may prove beneficial in future clinical applications, in particular with respect to a direct differentiation of stem cells in vitro.
The tremendous increase in primary and functional data brought about by the enhanced throughput of novel proteomics technologies clearly illustrates the urgent requirement for top-level bioinformatics support at all stages, from gathering and interpretation of primary data to their functional annotation and storage.
In silico prediction of functional protein interactions
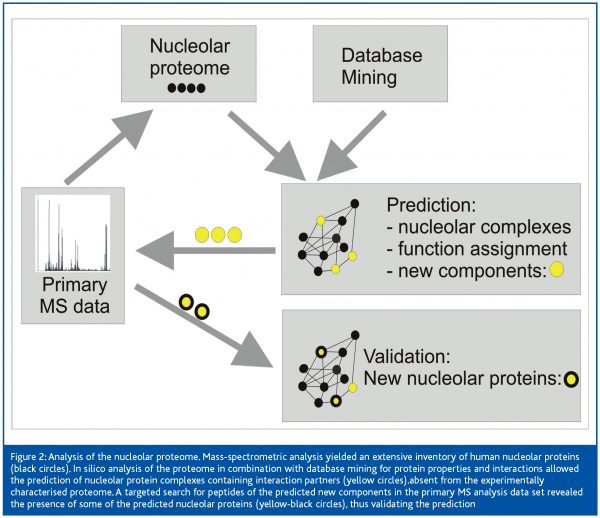
One example of the impact of fruitful collaborations between wet-lab and theoretical scientists within Interaction Proteome is the recent analysis of the nucleolar proteome4. An extensive inventory of human nucleolar proteins has recently been published by the proteomics experts in M. Mann’s group. Based on this compilation, the bioinformaticians around Soren Brunak, CBS, Technical University of Denmark, Lyngby, performed database-mining in various sources to predict functional protein complexes involving the identified nucleolar proteins. Neural nets were designed to use independently generated data – as, for example, a repeated demonstration of interaction by independent laboratories, or the presence of a nuclear translocation signal in the peptide sequence of interaction partners – to score the likelihood of the predicted complexes to exist and to be localised in the nucleolus. Thus combining primary information from various sources, a number of high-confidence functional nucleolar complexes was predicted. In a second step of reverse proteomics, the presence in the nucleolus of yet-unidentified components of these predicted complexes was postulated and these predictions were validated by a targeted search for corresponding peptides in the primary experimental data of the Mann group. The strategy is schematically depicted in Figure 2. This approach resulted in the experimental identification of 11 novel nucleolar proteins, along with the assignment of function to 49 previously uncharacterised proteins, and the identification of complexes serving various functions other than ribosome biogenesis. This investigation demonstrated how bioinformatics can support and extend high-throughput proteomics data collection and interpretation when joining forces with the experimentalists, and illustrates future ways forward in proteome research.
Data storage
The European Molecular-interactions data base MINT5 provides the central container for protein interaction data obtained in the project. The format of MINT has been adapted to the requirements of Interaction Proteome, so that the data sets can be directly used by the theoretical biologists for the modelling of protein interactions. In parallel, the curators at partner University of Rome, TorVergata efficiently integrate disparate data sets on protein interactions which are dispersed in the scientific literature into MINT. This allows the analysis of the newly discovered interactions in the context of the rich interaction data already deposited in the scientific literature by the community of traditional biologists. To combine these efforts with the other major interactions data bases world-wide, MINT was a founding member of the International Molecular Exchange Consortium (IMEx). This group of major public interaction data providers share curation efforts and exchange completed records on molecular interaction data, similar to successful global collaborations for protein and DNA sequences and for macromolecular structures. The five IMEx members draw upon different publications describing protein interactions and merge entries in a common dataset. The curation effort is minimised by collaboration.
The merged data are offered (unmodified) to the scientific community by the five database Web pages with slightly different style and analysis tools.
Outlook
With its Research Framework Programme 6, the European Commission devised new types of projects, or “instruments” – the Integrated Projects and Networks of Excellence – both characterised by multidisciplinarity and an increased size and budget as compared to earlier research projects. These project-types will be retained in only slightly modified form in the upcoming Research Framework Programme 7, which is due to start by the end of 2006. One major goal of these new instruments was the creation of the European Research Area via intensifying cross-discipline and cross-border collaborations in various fields of science.
What can we learn from Interaction Proteome in this respect? At project start, some collaborations between the individual partners already existed. During the first two years, interactions within the consortium intensified drastically, both with respect to intensity and multidisciplinarity. Rather than being enforced by the management structure, novel joint projects were initiated and state-of-the–art methods transferred between partner labs during informal discussions and exchanges during project workshops, meetings, lab visits – frequently between participants that had hardly known each other before hand. Although unforeseen to that extent, this development may, today, be regarded as one of the strongest aspects of the project. The collaboration within the multi-disciplinary consortium enables excellent progress in a multidisciplinary team, as outlined with some examples above. Furthermore, it provides a broad platform for the career development of young researchers within the consortium. Already, some of the members of Interaction Proteome have relocated within the consortium to initiate the next step in their career.
The performance of Interaction Proteome during its first two years of existence was scrutinised during the Mid-Term Review, initiated by the European Commission and performed with the assistance of two external experts, Prof. A. Emili, Toronto and Prof. D. Jones, London. In their evaluation report, the reviewers rated the project’s progress as excellent, in particular with regard to the multidisciplinary approaches taken, concluding with the remark that Interaction Proteome is “clearly a showcase project for EU research”.


References
- Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. (2005). Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 12, 2010-21.
- Van Damme P, Martens L, Van Damme J, Hugelier K, Staes A, Vandekerckhove J and Gevaert K. (2005). Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat Methods 2:771-7.
- Kratchmarova I, Blagoev B, Haack-Sorensen M, Kassem M, and Mann M. (2005). Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science 308(5727):14727.
- Hinsby AM, Kiemer L, Karlberg EO, Lage K, Fausboll A, Juncker AS, Andersen JS, Mann M, Brunak S (2006) A wiring of the human nucleolus.Mol Cell.22:285-95.
- http://mint.bio.uniroma2.it/mint




