Applications in target ID and validation
Posted: 11 November 2005 | | No comments yet
RNAi technology provides the ‘loss of function’ approach, which has been widely used in the last couple of years for analysis of gene function, and in drug discovery for identification and validation of potential drug target candidates. This technology is now widely applied for functional screens in order to identify disease associated signals and targets whose specific inhibition could potentially result in a curative effect for treatment of complex human diseases. The focus of such screens is evolving more towards druggable gene families rather than genome wide approaches – the latter being expensive, complex and sometimes confusing.
RNAi technology provides the ‘loss of function’ approach, which has been widely used in the last couple of years for analysis of gene function, and in drug discovery for identification and validation of potential drug target candidates. This technology is now widely applied for functional screens in order to identify disease associated signals and targets whose specific inhibition could potentially result in a curative effect for treatment of complex human diseases. The focus of such screens is evolving more towards druggable gene families rather than genome wide approaches – the latter being expensive, complex and sometimes confusing.
RNAi technology provides the ‘loss of function’ approach, which has been widely used in the last couple of years for analysis of gene function, and in drug discovery for identification and validation of potential drug target candidates. This technology is now widely applied for functional screens in order to identify disease associated signals and targets whose specific inhibition could potentially result in a curative effect for treatment of complex human diseases. The focus of such screens is evolving more towards druggable gene families rather than genome wide approaches – the latter being expensive, complex and sometimes confusing.
Modern technologies and resources such as the human genome project (sequence), gene chip experiments (gene expression profiling with microarrays chips) and advanced bioinformatics deliver a plethora of possible candidate targets for future drugs. Target validation – and particularly target invalidation – has become a crucial step in the drug discovery process. However, the availability of overwhelming amounts of genomic, transcriptomic and proteomic data has, paradoxically, made it difficult to select a molecular target for a full-scale drug development effort. The field of target validation promises to uncover the few real drug targets among the myriad ´omics´-candidates, and thus reduce costly attrition later in development1.
The challenge in target validation is to provide the usefulness of a specific target in vitro and ideally in vivo (animal model) as well. Correlation of a target’s presence with a disease relevant call or in vivo phenotype (e.g. through expression profiling) is not good enough; it is also essential to demonstrate that the target has a causal role in producing that phenotype. For a target to be selected for a drug development effort it must be expressed in the relevant cell type or tissue associated with human disease.
Modulation of the target’s activity in cell culture must ameliorate the relevant phenotype and have the same effect in an animal model of disease. Nevertheless, a target is not truly validated until a drug is proven effective in human trials.
Ideally, low molecular weight, target specific compounds can be used for validation but are often not available. One solution for dealing with the many targets, for which no agonists/antagonists are available, is modulating target protein function by blocking expression of corresponding mRNA2. Although there is no single universal solution for every target, there is a clear consensus among pharmaceutical companies that tools such as RNAi, antibodies and model organisms can be used to invalidate the vast majority of candidates identified through correlative target validation work3,4. RNAi compounds can then be administered in the original disease phenotype. A significant effect of such a compound on the disease phenotype indicates that the target in question could well play an important role in the disease itself.
RNA interference
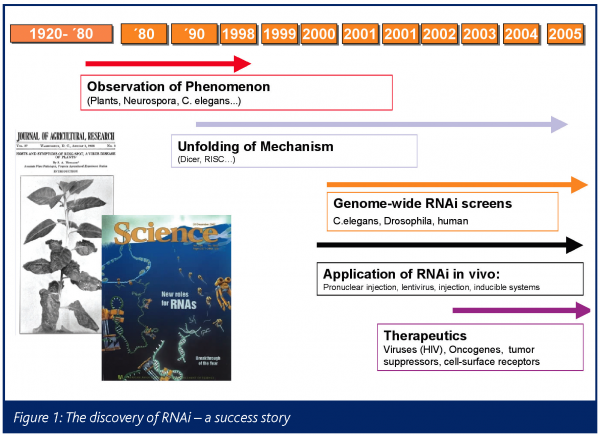
In 1998 the teams of Andy Fire and Craig Mello first realised that they could inactivate expression of specific genes in C.elegans by injecting the worms with double-stranded RNAs complimentary in sequence to the gene they wanted to inactivate5. In a series of elegant experiments it was demonstrated that double stranded RNA (dsRNA) induces posttranscriptional degradation of homologue transcripts entering a cellular pathway, that is commonly referred to as the RNA interference (RNAi), and has been observed in a variety of organisms including plant, fungi, insects, worms, protozoans and mammals (Figure 1)6-16.
The use of RNA interference for inhibiting gene expression represents a powerful tool in exploring gene function, identifying and validating new drug targets and treating disease. An additional important aspect of this validation approach is that it is flexible and scalable as, depending upon the bioassays and disease models available, RNAi compounds can be designed and selected for large numbers of drug target candidates. In numerous publications dose-response, longevity and correlation of protein and RNA depletion has been demonstrated with RNAi17-21.
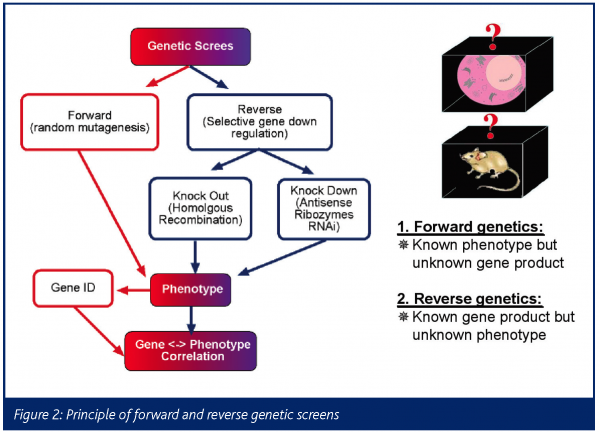
Reverse and forward genetic screens
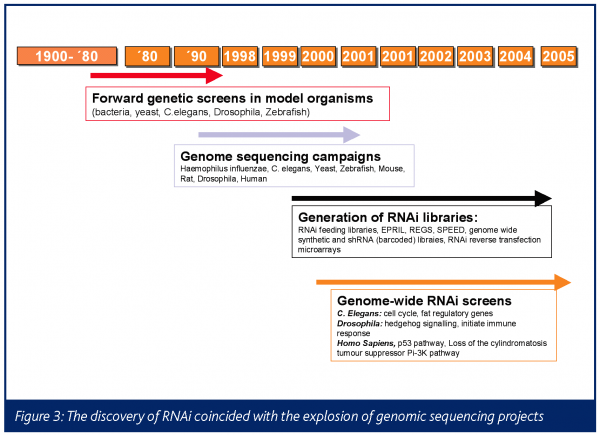
The traditional approach of forward genetics starts with a phenotype and then identifies the gene(s) or mutation that control or cause that trait (Figure 2). There have been many prominent examples for this approach in model organisms such as bacteria, yeast, C.elegans, Drosophila and Zebrafish22-25. In contrast is reverse genetics, in which one goes from a gene/sequence – often discovered via high-throughput sequencing and bioinformatics technologies – to its biological function26.
This is a direct result of the sequencing campaigns in Haemophilus influenzae, bacteria, yeasts, C. elegans, zebrafish, mouse, rat, Drosophila and human (Figure 3)27-30.
Reverse genetic methods are more amenable to whole genome high-throughput analysis than forward genetics. There is functional information for only ~15 per cent of human genes, at present. Loss-of-function genetic screens aim to identify gene function through inactivation of a gene (or its corresponding mRNA). Various technologies have been developed to study the effects of gene suppression in mammalian cells, including genetic suppressor elements (GSEs)31-34, antisense vectors35, ribozymes36, aptamer libraries37 and – more recently – RNA interference (RNAi)38-45. Through its use in large-scale screens and selection experiments RNAi is expected to provide great impact on cell biology.
Meanwhile, there are many examples of genome wide and focused siRNA screens such as in C. elegans (cell cycle, fat regulatory genes)46 Drosophila (hedgehog signalling, initiate immune response)47-48 and Homo sapiens (p53 pathway, loss of the cylindromatosis tumour suppressor Pi-3K pathway)49-51. These large-scale RNAi screens have already reproduced many previously hard-won genetic discoveries and have identified new genes that appear to function in well studied biological processes38,47,52 53.
Several academic and industrial research groups are following on the heels of this work to carry out more specialised focused RNAi screens. There are a number of examples for gene-family focused screens, especially in the field of kinases, as attractive potential pharmaceutical targets in cancer43,52,54,55,56,59. Drugs such as Gleevec and Iressa inhibit tumour growth by antagonising specific survival kinases, thus reversing the malignant phenotype. Such ‘targeted’ therapies offer enormous advantages over conventional broad-spectrum anti-cancer agents, which are frequently associated with non-specific toxic side effects. Large-scale RNAi screens can lead to the identification of human kinases that are essential for survival.
Critical success factors for high-throughput RNAi screens
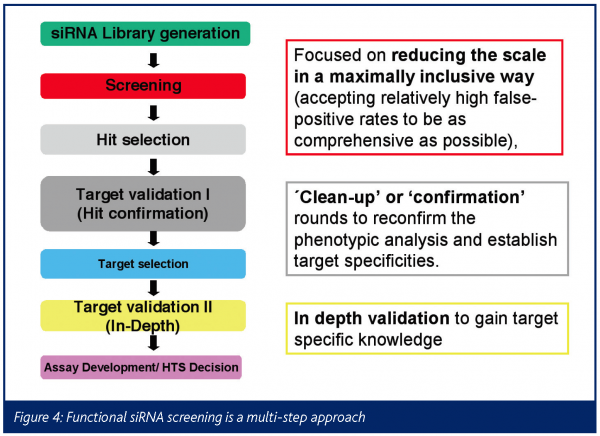
Effective functional siRNA screens represent a multi-pass approach. Several factors in the RNAi workflow and experimental design must be optimised to ensure the success of a high-throughput experiment, such as RNAi reagent selection, delivery methods, functional readouts, as well as collection, analysis and interpretation of data. The assay used for screening ultimately reflects the aim of the experiment, be it the study of genes in a particular pathway or the identification of genes involved in a disease process.
The primary screen reduces the scale of the library allowing it to be as comprehensive as possible, even at the expense of inclusion of false positive candidates. The initial positive gene candidates are prioritised based on how broad and prominent the effect is in different cell models. In a confirmation step putative targets are validated in a panel of different cellular models, assessing dose dependence and specificity tests e.g. the exclusion of off-target effects (the ‘clean-up step’). These experiments will increase the confidence of target specific effects against the off-target siRNA effects. Based on their functional characteristics and their known and/or predicted and experimentally validated biochemical functions (in-depth validation step), the newly identified candidate targets are prioritised for nomination and entry into the drug discovery process.
RNAi reagent selection
The siRNA sequence can significantly influence its potency (level of knockdown achieved) and specificity (stimulating phenotype through knockdown of the target gene only). Reconfirmation of gene annotation, sequence analysis of gene family homologues and splice variants, etc. is strongly recommended before designing the siRNAs by using one of many public or proprietary algorithms. Meanwhile there are multiple companies and academic resource centres in Europe and the US involved in the development and distribution of RNAi tools. These provide genome wide synthetic and shRNA (barcoded) libraries60, RNAi feeding libraries, EPRIL61, REGS62, SPEED63, or RNAi reverse transfection or adenoviral expression library microarrays, i.e arrayed-format libraries containing expression vectors that are deposited in an arrayed manner in defined locations64-66.
Choosing a model cell system
The choice of cell type should also be carefully considered for suitability with efficient, high-throughput siRNA transfection. In addition, some cell types may be more biologically responsive than others to the particular RNAi effect under study. The cell type chosen for RNAi experiments should be easy to transfect at high efficiencies and compatible with the downstream screening assay. It is advisable to perform initial RNAi experiments with more than one cell type, as there are numerous publications that show there to be highly significant differences between the level of responsiveness of different cell lines to knockdown of genes involved in the cell cycle43, 67.
siRNA delivery
High levels of gene knockdown are necessary for downstream analysis. This means that high transfection efficiency is also necessary. A reduction in transfection efficiency will reduce knockdown and will also reduce the level of phenotypic effects. This may make phenotypic effects difficult to detect and, hence, reduce reproducibility. Research suggests that using low siRNA concentrations in RNAi experiments lowers the risk of nonspecific effects68, 69. Dose-response experiments with independent duplexes are one of the best ways to confirm specificity, but are difficult to perform.
Confirmation of gene knockdown
Gene knockdown can be validated by various methods. The most widely used are quantitative real-time RT-PCR analysis at the mRNA level17 and western blot analysis at the protein level. The latter is restricted in its application for high-throughput analyses because antibodies for the protein of interest are not always available. Alternatively, a sense clone could be tagged to a GFP protein, co-transfected and subsequently the change of fluorescence measured by FACS analysis71, 72.
Control experiments
When setting up the experiment, it is vital to include adequate positive and negative controls to ensure that the high volume of data can be correctly interpreted and to account for variation in the different parameters of the procedure. Positive and negative (non-silencing) control siRNAs should be used in each experiment. Positive control siRNAs are siRNAs that are known to provide high knockdown of a target gene that produces the desired phenotype. Routine transfection of positive control siRNAs shows that transfection and assay conditions remain optimal. Small molecules or bioactive compounds that produce the phenotype under study can also be used as positive controls for assay conditions. For example, an apoptosis-inducing drug could be used as a positive control for an apoptosis screening assay. Alternatively, an inhibitory compound could be used to inhibit upstream components of the pathway under study, causing translocation of the protein examined in the screening. Non-silencing control siRNAs can be siRNAs with no homology to any known mammalian gene or siRNAs that are homologous to a gene that is not present in the cells under study (for example green fluorescent protein). Data from transfection of non-silencing siRNAs can be used to analyse the extent of nonspecific effects that may have occurred as a result of siRNA transfection. Phenotypic effects observed after knockdown must always be confirmed by one or more additional siRNAs targeted to different regions of the same mRNA.
Screening assays
The assays used for RNAi screens vary depending on the purpose of the experiment. Experiments can range from looking at a small group of gene targets for pathway analysis to screening the whole human genome for drug discovery. Assays may be carried out manually or can be partially or completely automated. The assay can range from a simple homogeneous cell-based assay to an assay that looks at changes in the sub-cellular distribution of a protein using a high-content automated confocal microscope70. An advantage of this type of research is that the assays don’t require high complexity to yield valuable information about cellular pathways and responses.
Management and mitigation of off-target effects
While siRNA mediated gene knockdown was originally purported to be highly specific, recent microarray studies have demonstrated RNAi-mediated ‘off-target’ gene modulation. Off-target effects can generate measurable phenotypes. siRNA have been identified that induce cell death in a target-independent, sequence-dependent manner83,84.
siRNA signatures are a sum of on- and off-target gene regulation73,78. The different gene silencing mechanisms of short interfering (si)RNAs and microRNAs present a challenge for siRNA design: on the one hand, mismatches between the mRNA target and the 3’ and central regions of the siRNA are usually detrimental for gene silencing; hence, an optimal siRNA should contain mismatches with other transcripts in these regions to avoid off-target effects. On the other hand, translational repression by microRNAs requires a near-perfect matching in the 5’ region, whereas a central bulge and partial binding in the 3’ region seem to be preferential for translational repression79-82.
Possible solutions to mitigate these artifacts are improved bioinformatics (through improving annotation of target sequence) and siRNA design to generate more efficient siRNAs. Chemical modifications, for example, to inactivate sense strand or increase stability (longer half life) and high-purity siRNA tools, can add significant specificity. From an experimental standpoint, by using the lowest possible siRNA concentration, siRNA pooling and siRNA redundancy, i.e. more than one siRNA/target, as well as careful monitoring of global cellular effects by for example expression analysis adds further confidence in the phenotypic results. Finally, rescue experiments (mutated target sequence, complementation by sense sequence) are one of the best biological proofs of specificity, however are technically very demanding85,86.
Decision support or how to choose targets wisely
The huge volumes and complex dependencies of data produced by such large-scale experiments make a clear workflow for data analysis processes mandatory, resulting in the development of novel data analysis strategies tailored to drug discovery.
Target selection requires the collection and analysis of a wealth of diverse information beyond DNA sequence and protein structure and role. Whereas initial target information is quite structured, it consists of a name and alternate names, DNA and protein sequences, protein structure, related genes and so forth, a target selection decision must be based on a wide range of largely unstructured information that is often derived from the published literature. Questions to be considered in selecting a target include the physiological and pathological role of the protein; diseases that the target could intervene; alternative approaches to achieving the same therapeutic aim; potential for side effects; patent and competitive landscape as well as the resources required.
The differences between structured and unstructured information present information management challenges, because it is not usually possible to rely on formal databases to manage all aspects of target selection. Summary annotation of candidate genes does not provide the necessary depth and context to support full assessment. They require powerful indexing and retrieval methods to ensure that collected information is accessible. Collecting and assessing the information needed to make a target-selection decision ideally requires the input of a wide range of experts from different functional areas in a company. Target selection should not be the exclusive province of bioinformaticians, but ideally ought to include input from chemistry, pathology, clinical development and even marketing and the legal department, and so forth. Criteria such as disease relevance, drugability, amenability to high throughput screening, feasibility of secondary test cascades, availability of reference compound, situation of intellectual property, freedom to operate, defined competition and so forth should provide a balanced target portfolio.
The future of RNAi screening in drug development
An additional value of RNAi screens is that they can be combined with other functional genomics assays, for example, transcriptional profiling and protein interaction experiments, resulting in more ‘-ome’ terminologies such as ‘phenome’, ‘transcriptome’, ‘interactome’87, 88. The future of genetic screening will require increasingly sophisticated assays both for conventional genetics as well as for RNAi-based approaches. Stained fat bodies Kaveh Ashrafi46, fluorescent reporter protein for genome instability Joris Polthof89, longevity and DNA repair89, 90 are just a few examples of such specialised genetic screens.
Summary
Target Discovery including target validation/invalidation is a crucial step in the early drug discovery process. RNAi is a powerful tool for target discovery facilitating the expedited mining of genomic information. Reverse loss-of-function screening such as RNAi-based high-throughput screens enable the discovery of proprietary targets for further drug development. Various published work demonstrated the successful application of siRNA screening. RNAi-based screening requires a highly defined workflow including automation, bioinformatics, multiple layers of control and a panel of robust assays. As with every technology that interferes with biological systems, there is the risk of artifacts (false positives and false negatives). With reasonable efforts, these can be managed and mitigated to a certain extent. Several selection criteria can play an important role in determining reliable target priority, for example target tissue expression profiles, degree and quality of available intellectual property, drugability and access to necessary molecular validation tools – all of which can help focus discovery efforts on the most promising drug target candidates. Once the target validation has been completed, the full drug discovery capability of a pharmaceutical company can be focused with confidence on identification and development of drugs specific to the confirmed target.
Acknowledgements
Thanks to Dmitry Samarsky from Dharmacon for his critical review of this article.




References
- Szymkowski, D.E., Target validation joins the pharma fold. TARGETS, 2003. 2(#1 February 2003): p. 8-10.
- Stephan, J.P., From functional genomics to target validation: the quest for better drugs. Drug Discov Today, 2004. 3(1): p. 7-9.
- Chopra, M., et al., Using RNA interference to modulate gene expression. TARGETS, 2003. 1(3): p. 102-108.
- Constans, A., RNAi for the Masses. The Scientist, 2002. 16(9): p. 36.
- Fire, A., et al., Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 1998. 391(6669): p. 806-11.
- van der Krol, A.R., et al., Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell, 1990. 2(4): p. 291-9.
- Mol, J.N., et al., Regulation of plant gene expression by antisense RNA. FEBS Lett, 1990. 268(2): p. 427-30.
- Napoli, C., C. Lemieux, and R. Jorgensen, Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. Plant Cell, 1990. 2(4): p. 279-289.
- Romano, N., et al., Quelling: transient inactivation of gene in Neurospora crassa by transformation with homologous sequences. Mol Microbiol, 1992. 6(22): p. 3343-53.
- Silva, J., et al., RNA-interference-based functional genomics in mammalian cells: reverse genetics coming of age. Oncogene, 2004. 23(51): p. 8401-9.
- Paddison, P.J., et al., Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev, 2002. 16(8): p. 948-58.
- Ketting, R.F., et al., Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev, 2001. 15(20): p. 2654-9.
- Bernstein, E., A.M. Denli, and G.J. Hannon, The rest is silence. Rna, 2001. 7(11): p. 1509-21.
- Elbashir, S.M., et al., Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 2001. 411(6836): p. 494-8.
- Elbashir, S.M., et al., Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods, 2002. 26(2): p. 199-213.
- Sijen, T., et al., On the role of RNA amplification in dsRNA-triggered gene silencing. Cell, 2001. 107(4): p. 465-76.
- Scherer, L., et al., RNAi applications in mammalian cells. Biotechniques, 2004. 36(4): p. 557-61.
- Paul, C.P., et al., Effective expression of small interfering RNA in human cells. Nat Biotechnol, 2002. 20(5): p. 505-8.
- Paddison, P.J. and G.J. Hannon, RNA interference: the new somatic cell genetics? Cancer Cell, 2002. 2(1): p. 17-23.
- Dykxhoorn, D.M., C.D. Novina, and P.A. Sharp, Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol, 2003. 4(6): p. 457-67.
- Novina, C.D. and P.A. Sharp, The RNAi revolution. Nature, 2004. 430(6996): p. 161-4.
- Stark, G.R. and A.V. Gudkov, Forward genetics in mammalian cells: functional approaches to gene discovery. Hum Mol Genet, 1999. 8(10): p. 1925-38.
- St Johnston, D., The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet, 2002. 3(3): p. 176-88.
- King, D.P. and J.S. Takahashi, Forward genetic approaches to circadian clocks in mice. Cold Spring Harb Symp Quant Biol, 1996. 61: p. 295-302.
- Pickart, M.A., et al., Functional genomics tools for the analysis of zebrafish pigment. Pigment Cell Res, 2004. 17(5): p. 461-70.
- Adams, M.D. and J.J. Sekelsky, From sequence to phenotype: reverse genetics in Drosophila melanogaster. Nat Rev Genet, 2002. 3(3): p. 189-98.
- Adams, M.D., et al., The genome sequence of Drosophila melanogaster. Science, 2000. 287(5461): p. 2185-95.
- Carninci, P., et al., The transcriptional landscape of the mammalian genome. Science, 2005. 309(5740): p. 1559-63.
- Gibbs, R.A., et al., Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature, 2004. 428(6982): p. 493-521.
- Venter, J.C., et al., The sequence of the human genome. Science, 2001. 291(5507): p. 1304-51.
- Gudkov, A.V., et al., Isolation of genetic suppressor elements, inducing resistance to topoisomerase II-interactive cytotoxic drugs, from human topoisomerase II cDNA. Proceedings of the National Academy of Science of the USA, 1993. 90: p. 3231-3235.
- Gudkov, A.V., et al., Cloning mammalian genes by expression selction of gentic suppressor elements: association of kinesin with drug resistance and cell immortalization. Proceedings of the National Academy of Science of the USA, 1994. 91: p. 3722-3748.
- Gudkov, A.V. and I.B. Ronnison, Isolation of genetic supressor elements (gseS) from random fragented cDNA libraries in retorviral vectors. Protocols for cDAN libraries, ed. G. Cowell and A.C. A. 1998: Humana Press.
- Roninson, I.B., et al., Genetic suppressor elements: new tools for molecular oncology-thirteenth cornelius P . Rhoads memorial award lecture. Cancer Research, 1995. 55: p. 4023-4028.
- Kimchi, A., Antisense libraries to isolate tumor suppressor genes. Methods Mol Biol, 2003. 222: p. 399-412.
- Beger, C., et al., Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc Natl Acad Sci U S A, 2001. 98(1): p. 130-5.
- Colas, P., et al., Genetic selection of peptide aptamers that recognize and inhibit cyclin-dependent kinase 2. Nature, 1996. 380(6574): p. 548-50.
- Kleino, A., et al., Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. Embo J, 2005. 15: p. 15.
- Muller, P., et al., Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature, 2005. 436(7052): p. 871-5.
- Baeg, G.H., R. Zhou, and N. Perrimon, Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev, 2005. 19(16): p. 1861-70. Epub 2005 Jul 29.
- Westbrook, T.F., et al., A genetic screen for candidate tumor suppressors identifies REST. Cell, 2005. 121(6): p. 837-48.
- Bernards, R., A functional approach to questions about life, death, and phosphorylation. Cancer Cell, 2005. 7(6): p. 503-4.
- MacKeigan, J.P., L.O. Murphy, and J. Blenis, Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol, 2005. 7(6): p. 591-600. Epub 2005 May 1.
- Boutros, M., et al., Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science, 2004. 303(5659): p. 832-5.
- Tewari, M. and M. Vidal, RNAi on the apoptosis TRAIL: the mammalian cell genetic screen comes of age. Dev Cell, 2003. 5(4): p. 534-5.
- Ashrafi, K., et al., Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature, 2003. 421(6920): p. 268-72.
- Foley, E. and P.H. O’Farrell, Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol, 2004. 2(8): p. E203. Epub 2004 Jun 22.
- Panakova, D., et al., Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature, 2005. 435(7038): p. 58-65.
- Brummelkamp, T.R., et al., Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature, 2003. 424(6950): p. 797-801.
- Hudson, J.D., et al., A proinflammatory cytokine inhibits p53 tumor suppressor activity. J Exp Med, 1999. 190(10): p. 1375-82.
- Carnero, A., et al., Loss-of-function genetics in mammalian cells: the p53 tumor suppressor model. Nucleic Acids Res, 2000. 28(11): p. 2234-41.
- Berns, K., et al., A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature, 2004. 428(6981): p. 431-7.
- DasGupta, R., et al., Functional genomic analysis of the Wnt-wingless signaling pathway. Science, 2005. 308(5723): p. 826-33. Epub 2005 Apr 7.
- Shelton, J.G., et al., The epidermal growth factor receptor gene family as a target for therapeutic intervention in numerous cancers: what’s genetics got to do with it? Expert Opin Ther Targets, 2005. 9(5): p. 1009-30.
- Rabindran, S.K., Antitumor activity of HER-2 inhibitors. Cancer Lett, 2005. 227(1): p. 9-23. Epub 2004 Dec 15.
- Dietrich, S., et al., Role of c-MET in upper aerodigestive malignancies – from biology to novel therapies. J Environ Pathol Toxicol Oncol, 2005. 24(3): p. 149-62.
- Corso, S., P.M. Comoglio, and S. Giordano, Cancer therapy: can the challenge be MET? Trends Mol Med, 2005. 11(6): p. 284-92.
- Silva, C.M., Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis. Oncogene, 2004. 23(48): p. 8017-23.
- Ward, J.P., et al., Protein kinases in vascular smooth muscle tone–role in the pulmonary vasculature and hypoxic pulmonary vasoconstriction. Pharmacol Ther, 2004. 104(3): p. 207-31.
- Brummelkamp, T.R. and R. Bernards, New tools for functional mammalian cancer genetics. Nat Rev Cancer, 2003. 3(10): p. 781-9.
- Shirane, D., et al., Enzymatic production of RNAi libraries from cDNAs. Nat Genet, 2004. 36(2): p. 190-6.
- Sen, G., et al., Restriction enzyme-generated siRNA (REGS) vectors and libraries. Nat Genet, 2004. 36(2): p. 183-9.
- Luo, B., A.D. Heard, and H.F. Lodish, Small interfering RNA production by enzymatic engineering of DNA (SPEED). Proc Natl Acad Sci U S A, 2004. 101(15): p. 5494-9.
- Michiels, F., et al., Arrayed adenoviral expression libraries for functional screening. Nat Biotechnol, 2002. 20(11): p. 1154-7. Epub 2002 Sep 30.
- Arts, G.J., et al., Adenoviral vectors expressing siRNAs for discovery and validation of gene function. Genome Res, 2003. 13(10): p. 2325-32. Epub 2003 Sep 15.
- Williams, N.S., et al., Identification and validation of genes involved in the pathogenesis of colorectal cancer using cDNA microarrays and RNA interference. Clin Cancer Res, 2003. 9(3): p. 931-46.
- Hahn, P., et al., A genomewide perspective of gene expression – integration of QIAGEN RNAi technologies and Affymetrix GeneChip Arrays. QIAGEN News, 2005. e8.
- Semizarov, D., et al., Specificity of short interfering RNA determined through gene expression signatures. Proc Natl Acad Sci U S A, 2003. 100(11): p. 6347-52. Epub 2003 May 13.
- Persengiev, S.P., X. Zhu, and M.R. Green, Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs). Rna, 2004. 10(1): p. 12-8.
- Bertelsen, M. and A. Sanfridson, Inflammatory pathway analysis using a high content screening platform. Assay Drug Dev Technol, 2005. 3(3): p. 261-71.
- Sui, G. and Y. Shi, Gene silencing by a DNA vector-based RNAi technology. Methods Mol Biol, 2005. 309: p. 205-18.
- Harper, S.Q. and B.L. Davidson, Plasmid-based RNA interference: construction of small-hairpin RNA expression vectors. Methods Mol Biol, 2005. 309: p. 219-35.
- Achenbach, T.V., B. Brunner, and K. Heermeier, Oligonucleotide-based knockdown technologies: antisense versus RNA interference. Chembiochem, 2003. 4(10): p. 928-35.
- Scherer, L.J. and J.J. Rossi, Approaches for the sequence-specific knockdown of mRNA. Nat Biotechnol, 2003. 21(12): p. 1457-65.
- Miyagishi, M., et al., Optimization of an siRNA-expression system with an improved hairpin and its significant suppressive effects in mammalian cells. J Gene Med, 2004. 6(7): p. 715-23.
- Duxbury, M.S. and E.E. Whang, RNA interference: a practical approach. J Surg Res, 2004. 117(2): p. 339-44.
- Sledz, C.A. and B.R. Williams, RNA interference and double-stranded-RNA-activated pathways. Biochem Soc Trans, 2004. 32(Pt 6): p. 952-6.
- Scherr, M. and M. Eder, RNAi in functional genomics. Curr Opin Mol Ther, 2004. 6(2): p. 129-35.
- Scacheri, P.C., et al., Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci U S A, 2004. 101(7): p. 1892-7. Epub 2004 Feb 9.
- Silva, J.M., et al., RNA interference microarrays: high-throughput loss-of-function genetics in mammalian cells. Proc Natl Acad Sci U S A, 2004. 101(17): p. 6548-52.
- Snove, O., Jr. and T. Holen, Many commonly used siRNAs risk off-target activity. Biochem Biophys Res Commun, 2004. 319(1): p. 256-63.
- Amarzguioui, M., et al., Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res, 2003. 31(2): p. 589-95.
- Jackson, A.L., et al., Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol, 2003. 21(6): p. 635-7.
- Chi, J.T., et al., Genomewide view of gene silencing by small interfering RNAs. Proc Natl Acad Sci U S A, 2003. 2: p. 2.
- Sarov, M. and A.F. Stewart, The best control for the specificity of RNAi. Trends Biotechnol, 2005. 23(9): p. 446-8.
- Du, Q., et al., A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res, 2005. 33(5): p. 1671-7. Print 2005.
- Walhout, A.J., et al., Integrating interactome, phenome, and transcriptome mapping data for the C. elegans germline. Curr Biol, 2002. 12(22): p. 1952-8.
- Davy, A., et al., A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep, 2001. 2(9): p. 821-8.
- Pothof, J., et al., Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev, 2003. 17(4): p. 443-8.
- Kamath, R.S., et al., Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature, 2003. 421(6920): p. 231-7.




