The promise and pitfalls
Posted: 22 August 2005 | | No comments yet
Perhaps the most significant technological advancement in the study of gene function in the post-genome era has been the discovery that RNA interference (RNAi) can be exploited for depletion of endogenous mRNA in mammalian cells. As the pharmaceutical industry has fallen under intense pressure to both identify and validate high-quality drug targets, the lure of bona fide genome-wide functional analysis and target identification using small interfering RNA (siRNA) has fueled the interest in what can now be truly called ‘functional’ genomics.
Perhaps the most significant technological advancement in the study of gene function in the post-genome era has been the discovery that RNA interference (RNAi) can be exploited for depletion of endogenous mRNA in mammalian cells. As the pharmaceutical industry has fallen under intense pressure to both identify and validate high-quality drug targets, the lure of bona fide genome-wide functional analysis and target identification using small interfering RNA (siRNA) has fueled the interest in what can now be truly called ‘functional’ genomics.
Perhaps the most significant technological advancement in the study of gene function in the post-genome era has been the discovery that RNA interference (RNAi) can be exploited for depletion of endogenous mRNA in mammalian cells. As the pharmaceutical industry has fallen under intense pressure to both identify and validate high-quality drug targets, the lure of bona fide genome-wide functional analysis and target identification using small interfering RNA (siRNA) has fueled the interest in what can now be truly called ‘functional’ genomics.
RNA interference (RNAi) refers to the process by which short double-stranded RNA molecules enter into a enzymatic complex known as RISC (RNA-induced silencing complex) and induce sequence-specific degradation of a target mRNAs. In 1998, Fire and colleagues1 first demonstrated that RNAi could be induced in a model organism by injection of in vitro transcribed long double-stranded RNA into Caenorhabditis elegans. The observation that RNAi could be recapitulated in a model organism provided evidence that post-transcriptional gene silencing previously observed in plants and fungi were most likely also due to RNA interference. Three years later, Elbashir and colleagues2 reported the first successful application of RNAi in mammalian cells using 21-nucleotide RNA duplexes, which had previously been shown to be the active component of RNAi in Drosophila embryos3. It is now understood that long double stranded RNA molecules are enzymatically cleaved by an evolutionarily conserved RNase III activity known as DICER, yielding the active 21-25 base pair dsRNA products. Such small synthetic dsRNAs do not appear to induce a potent interferon response and are relatively stable, making them ideal tools for elucidation of gene function.
Neither the impact nor the potential of these collective discoveries can be overstated. RNAi has become a standard approach to identify functions of genes in a variety of in vitro and in vivo model systems. After a handful of initial manuscripts appeared in 2001, more than 2000 papers describing RNAi were published in 2004. Armed with only the predicted sequences of putative genes, a researcher can rapidly design gene-specific antagonists to probe the roles of these genes in a variety of cellular processes and physiological settings. Numerous recent reviews have highlighted the mechanism of RNAi4,5, and the considerations for design of an interpretable RNAi experiment6. However, as the genomes of many organisms have been fully sequenced, it is both technically and economically feasible to produce RNAi reagents targeting the complete predicted transcriptome of an organism for systematic, high-throughput analysis of gene function. Such large-scale screening programs present several unique challenges with regard to experimental design and interpretation, especially in light of the potential consequence of off-target effects.
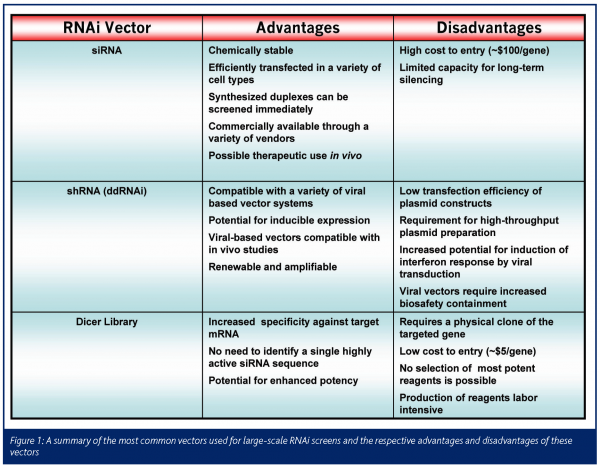
RNAi reagent selection
To date, two main strategies have been utilized for RNAi experimentation in mammalian cells: siRNA and shRNA. siRNA (small interfering RNA) traditionally refers to a double-stranded RNA oligonucleotide created by the annealing of two complementary, chemically synthesised single strands of RNA. ShRNA (short-hairpin RNA), first described by Brummelkamp and colleagues7, describes a precursor molecule of ~70 nucleotides which consists of reverse complimentary ~21-base pair sequences separated by a 6-10 base pair linker sequence. Once transcribed, this sequence folds back upon itself to form a hairpin structure. The single-stranded loop is recognised and processed by DICER to yield a mature 21-base pair siRNA that enters into the RISC. This hairpin is normally generated via a DNA vector and as such this approach has been termed ddRNA (DNA-directed RNAi). Both of these strategies have advantages and disadvantages for use in large-scale screens. Synthetic siRNA, while relatively costly, are remarkably stable and can be utilised for screening immediately upon synthesis. Unlike DNA constructs, for which transfection efficiency can vary significantly between cell lines, siRNA are efficiently transfected into a variety of transformed and primary cell types. shRNA contructs are relatively inexpensive, as the DNA cloning can be accomplished in high density format. However, the preparation of large-scale arrayed ddRNAi collections requires expertise in high-throughput, high-quality DNA plasmid preparation. An additional advantage of ddRNAi is the ability to utilise various viral-based vector systems, such as retroviruses8 and adenoviruses9, each of which assure high efficiency transfer, broad cell type tropism and the potential for in vivo experimentation. In addition, lenti-, retro- and adeno-associated viral vector systems can give stable expression of shRNA constructs. Kittler et al10. described an alternative approach to produce RNAi reagents in which a cDNA is bidirectionally in vitro transcribed to generate a long, double stranded RNA which is then subject to digestion by RNAse III processing. This process yields a mixture of random siRNAs for a given gene, thus eliminating the need for identification of exquisitely specific or potent individual siRNA against each targeted gene. Such an approach may mitigate off target effects due to the weak complimentarity of a single siRNA to other genes.
Assay design
In theory, RNAi screening for the elucidation of gene function is similar to screening small molecule chemical libraries in that the key to success is the development of robust, biologically relevant cell-based assays. Indeed, the time and cost associated with the development, optimisation and performance of a single screen pales in comparison to the time and cost associated with the biological validation of the screening results. To date, two basic strategies have been utilised for large-scale screens: multiwell arrays of RNAi reagents targeting single genes and complex pools of RNAi reagents. While each of these approaches has certain advantages, the ultimate choice of assay format is dictated by considerations specific to the relevant biological question being addressed. In the first large-scale mammalian RNAi screen, Brummelkamp and colleagues11 utilised a shRNA vector library targeting 50 genes in the de-ubiquitinating enzyme (DUB) family. An arrayed pool of four shRNA vectors targeting each DUB gene was transfected into U2-OS cells and the enhancement of tumor necrosis factor-α (TNF- α)-activation of an NF-κB-responsive reporter construct was assayed. This strategy ultimately led to the identification of CYLD as a factor capable of de-ubiquinating TRAF2, a key signaling component in the NF-κB pathway. Shortly thereafter, Aza-Blanc and colleagues12 described the use of a large-scale arrayed siRNA library targeting more than 500 individual genes to identify regulators of TRAIL-induced apoptosis in HeLa. These two reports heralded a fundamental change in the paradigm of genome analysis. Suddenly, genome-wide RNAi screens in mammalian cells were not just possible, but inevitable. A subsequent report by Berns et al.13 raised the bar higher still. In this study, a complex pool of shRNA-encoding vectors targeting nearly 8,000 genes was used to identify modulators of p53-dependent proliferation arrest. As the putative active shRNAs emerging from this screen stimulated clonal cell proliferation, genomic DNA from rescued cell clones was analysed to identify active shRNA sequences.
Off-target effects
The term ‘off-target effect‘ has been used to describe a variety of effects of siRNAs that are independent of the silencing of the intended mRNA target, which can ultimately confound the interpretation of RNAi experiments. These include the specific silencing of a gene other than the intended target via an siRNA or miRNA effect as well as the non-specific induction of the cellular interferon response. The extent to which individual siRNA duplexes are capable of efficiently silencing an mRNA demonstrating imperfect complementarity has been addressed in several well-designed studies, albeit with surprisingly different conclusions. Chi and colleagues14 addressed this question by introducing an siRNA specific for the enhanced green fluorescent protein (eGFP) of Aequoria victoria or a scrambled control siRNA into HEK293 that had been either transiently or stably transfected with an eGFP expression vector. These cells were then subject to expression profiling via DNA microarrays and, surprisingly, the authors observed no significant changes in expression of the approximately 20,000 detectable cellular RNAs. This suggests that, for at least the siRNA sequences used in this study, significant off-target effects were not observed. Also utilising expression profiling, Semizarov and colleagues15 came to a similar conclusion examining independent siRNA directed against three genes, AKT1, Plk1, and Rb into H1299 cells. At low concentrations (5 and 20nM), multiple siRNAs against the target genes yielded similar patterns of gene modulation that were specific to the depletion of the intended target mRNA. However, at higher concentrations (100nM), strong induction of a common gene set by all siRNAs was observed. This data suggests that off-target effects increase with siRNA concentration, arguing for the use of the lowest possible active siRNA concentration for loss-of-function studies. A more sobering view of the impact of off-target effects was presented by Jackson and colleagues16. In this report, off-target effects were shown to be distinctively siRNA- and not target-specific and the authors conclude that 15 nucleotides and perhaps as little as 11 contiguous base pairs, are sufficient to induce silencing by an siRNA duplex. The potential impact of these observations on large-scale RNAi screening is cause for concern. As siRNA are designed to maximise specificity for their intended target mRNA (and that specificity is calculated for the entire 21 base pair sequence), numerous ’off-target‘ genes may be effectively silenced by any given siRNA. So, as individual siRNA are likely to have a relatively unique spectrum of off-target effects, it stands to reason that genome-scale siRNA screens will yield a significant percentage of reproducible hits which are not necessarily attributable to the depletion of the intended target mRNA. However, independent siRNAs targeting the same transcript should not display identical off target effects. This supports the argument that multiple independent siRNAs should be utilised in the execution of large-scale screens and the probability of identifying multiple independent active siRNA directed against a single intended target will increase proportionately to the number of gene-specific siRNAs screened.
A second category of off-target effect that is of concern for large-scale screening, is the potential microRNA (miRNA) effect of siRNA duplexes. miRNAs are a class of naturally occurring double-stranded RNA molecules that are processed from a longer imperfect hairpin structure to create a pre-miRNA of approximately 70 nucleotides17. This pre-miRNA is then processed by the same cellular machinery that processes siRNA, resulting in a functional, single stranded molecule. The biochemical mechanism of target inhibition by miRNA is distinct from that of siRNAs. miRNAs are thought to bind to target mRNAs with some degree of imperfect complementarity and subsequently block efficient translation of the cognate protein in a RISC-independent process. While it was first believed that the functional consequences of siRNA and miRNA were exclusive, several recent reports have described the miRNA-activities of siRNA duplexes18,19. Because the sequence requirements are less stringent for miRNA function than for siRNA function, there is a potential that a single siRNA could result in translational repression of multiple off-target mRNAs. Currently it is not clear if single or multiple binding sites are required for an siRNA to show miRNA activity and thus it is not clear how off-target miRNA effects will impact siRNA screens.
Finally, it was initially proposed that the active 19-21 base pair sequence of an siRNA was insufficiently long to induce the innate double-stranded RNA response in transfected cells. This response, thought to be a conserved mechanism of resistance to RNA viruses, is mediated by activation of the dsRNA-dependent protein kinase, PKR and results in the upregulation of a variety of interferon-responsive genes. Several recent reports suggest that not only do some viral-based delivery vectors induce an interferon response, but also that the siRNA sequences themselves are capable of doing so in a sequence-specific manner20,21. Clearly, further investigation is warranted to determine the potential for interferon response induced by siRNA and this presents a potential hurdle for the use of RNAi-based therapeutics in man. Ultimately, the potential off-target effects of siRNA will be a major consideration for the interpretation of large-scale screening efforts. Ideally, a primary screen and secondary verification studies should utilise multiple validated RNAi reagents against a given target mRNA. Due to the labour and cost associated with such an effort, a more economical alternative is to use rational design strategies to identify general rules which result in the enhanced potency and specificity of siRNA.
Rational siRNA design
Shortly after the first demonstration of RNA interference initiated by 21-nucleotide dsRNAs, general rules for rational siRNA design began to emerge. The initial studies of siRNA in mammalian cells were performed with siRNA duplexes characterised by symmetric 2-nucleotide (primarily thymidine) overhanging 3’ ends2. Additional details followed shortly thereafter, including considerations for GC content (approximately 50%) and position with target mRNA (50-100 nucleotides downstream of the start codon)22. Two concurrently published studies point to an additional factor guiding siRNA efficacy: the so-called strand bias of incorporation into the RISC. As either of two strands of an siRNA duplex can be theoretically incorporated into the RISC, understanding the rules that guide this selection can greatly enhance the potential for the functional antisense strand (with respect to the intended target) to be preferentially incorporated. Using a novel RISC-capture assay, Schwarz and colleagues23 demonstrated that the strand of an siRNA that contains a less stable 5’ end was preferentially incorporated into the RISC. Khvorova and colleagues24 arrived at a similar conclusion via an entirely distinct approach. By observing the thermodynamic profiles of nearly 300 naturally occurring miRNAs from various species, a significant bias towards unstable 5’ ends was observed. In addition, an unstable 5’ end strongly correlated with potent siRNAs while an unstable 3’ end strongly correlated with an inactive one. These studies taken together demonstrate that subtle differences in thermodynamic stability may play a major role in the ’on-target‘ silencing potential of any given siRNA. The ’art‘ of predicting specific and potent siRNA will ultimately be aided by the development of algorthims based upon neural networks, as has been previously demonstrated for antisense oligonucleotides (ASO)25.
Conclusion
Large-scale screening with RNAi reagent collections holds tremendous promise for the identification of high potential drug targets. Using siRNAs as model antagonists, it should be possible to identify which of the 5,000 to 10,000 potential drug targets in the human genome26 are worth pursuing as therapeutic targets. While several successful mammalian RNAi screens have been reported in the past two years, it is important to recognise that screening results represent only the initial step in the elucidation of individual gene functions. Rigorous validation of screening results, including verification with independent siRNAs, correlation to protein depletion and phenotypic rescue with an untargetable cDNA construct6,27, will guarantee accurate assessment of functional activity. Ultimately, understanding the limitations of RNAi technology represents a critical factor in the translation of large-scale screening from a promising technology to increased efficiency in the pharmaceutical industry.

References
- Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature (1998), 391, 806-811.
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature (2001) 411, 494-498.
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. RNA interference is mediated by 21- an 22-nucleotide RNAs. Genes Dev (2001), 15, 188-200.
- Dykxhoorn, D., Novina, C., and Sharp, P. Killing the messenger: Shorts RNAs that silence gene expression. Nature Rev. Mol. Cell Biol. (2003) 4,457-467.
- Dorsett, Y. and Tuschl, T. siRNAs: Applications in functional genomics and potential as therapeutics. Nature Rev. Drug Disc. (2004) 3, 318-329.
- Huppi, K., Martin, S.E., and Caplen, N.J. Defining and assaying RNAi in mammalian cells. Mol. Cell (2005), 17, 1-10.
- Brummelkamp, T. R., Bernards, R., and Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science (2002), 296, 550-553.
- Paddison, P. J., Silva, J. M., et. al. A resource for large-scale RNA-interference-based screens in mammals. Nature (2004), 428, 427-431.
- Arts, G., Langemeijer, E., et. al. Adenoviral vectors expressing siRNAs for discovery and validation of gene function. Genome Res. (2003), 13, 2325-2332.
- Kittler, R., Putz, G., et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature (2004), 432, 1036-1040.
- Brummelkamp, T. R., Nijman, S. M., Dirac, A. M., and Bernards, R. Loss of the cylindomatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature (2003), 424, 797-801.
- Aza-Blanc, P., Cooper, C. L., Wagner, K., Batalov, S., Deveraux, Q. L., and Cooke, M. P. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol. Cell (2003), 12, 627-637.
- Berns, K. Hijmans, E. M., et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature (2004), 32, 893-901.
- Chi, J. T., Chang, H. Y., Wang, N. N., Chang, D. S., Dunphy, N., and Brown, P. O. Genomewide view of gene silencing by small intergering RNAs. Proc. Nat. Acad. Sci. (2003), 100, 6343-6346.
- Semizarov, D., Frost, L., Sarthy, A., Kroeger, P., Halbert, D. N., and Fesik, S. W. Specificity of short interfering RNA determined through gene expression signatures. Proc. Nat. Acad. Sci. (2003), 100, 6347-6352.
- Jackson, A. L., Bartz, S. R., Schelter, J., Kobayashi, A. V., Burchard, J., Mao, M., Li, B., Cavet, G., and Linsley, P. S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. (2003), 21, 635-637.
- Bartel, D. P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell (2004), 116, 281-297.
- Scacheri, P. C., Rozenblatt-Rosen, O., et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Nat. Acad. Sci. (2004), 101, 1892-1897.
- Doench, J. G., Peterson, C. P., and Sharp, P. A. siRNAs can function as miRNAs. Genes Dev. (2003), 17, 438-442.
- Sledz, C. A., Holko, M., de Veer, M. J., Silverman, R. H., and Williams, B. R. G. Activation of the interferon system by short-interfering RNAs. Nature Cell Biol. (2003), 5, 834-839.
- Bridge, A. J., Pebernard, S., Ducraux, A., Nicoulaz, N., and Iggo, R. Induction of an interferon response by RNAi vectors in mammalian cells. Nature Genet. (2003), 34, 263-264.
- Tuschl, T., Elbashir, S., Harborth, J., and Weber, K. The siRNA user guide. Published online at http://www.rockefeller.edu/labheads/tuschl/sirna.html
- Schwarz, D. S., Hutvagner, G., Du, T., Xu, X., Aronin, N., and Zamore, P. Asymmetry in the assembly of the RNAi enzyme complex. Cell (2003), 115, 199-208.
- Khvorova, A., Reynolds, A., and Jayasena, S. D. Functional siRNAs and miRNAs exhibit strand bias. Cell (2003), 115, 209-216.
- Giddings, M. C., Shah, A. A., Freier, S., Atkins, J. F., Gesteland, R. F., and Matveeva, O. V. Artificial neural network prediction of antisense oligodeoxynucleotides. Nuc. Acids Res. (2002), 30, 4295-4304.
- Drews, J. Biotechnology’s metamorphosis into a drug discovery industry. Nature Biotech. (1998), 16 (Suppl), 22-24.
- Whither RNAi? Nature Cell Biol. (2003), 5, 489-490.




