Plasma protein biomarker discovery
Posted: 22 August 2005 | | No comments yet
Less than a week after Nature and Science published the special issues on the ’blueprint‘ for the human genome sequence 15-16 Feb, 2001, the Financial Times of 21 February, 2001, ran a major article about proteomics, calling proteins “the real stuff of life”. Proteins are, indeed, the effector molecules for most cellular actions and interactions. As attention has migrated from genome sequences to genetic variation and functional genomics, proteomics has gradually emerged as a potentially powerful set of technologies for biomarker discovery and mechanistic studies important to drug development and drug safety surveillance1-4.
Less than a week after Nature and Science published the special issues on the ’blueprint‘ for the human genome sequence 15-16 Feb, 2001, the Financial Times of 21 February, 2001, ran a major article about proteomics, calling proteins “the real stuff of life”. Proteins are, indeed, the effector molecules for most cellular actions and interactions. As attention has migrated from genome sequences to genetic variation and functional genomics, proteomics has gradually emerged as a potentially powerful set of technologies for biomarker discovery and mechanistic studies important to drug development and drug safety surveillance1-4.
Less than a week after Nature and Science published the special issues on the ’blueprint‘ for the human genome sequence 15-16 Feb, 2001, the Financial Times of 21 February, 2001, ran a major article about proteomics, calling proteins “the real stuff of life”. Proteins are, indeed, the effector molecules for most cellular actions and interactions. As attention has migrated from genome sequences to genetic variation and functional genomics, proteomics has gradually emerged as a potentially powerful set of technologies for biomarker discovery and mechanistic studies important to drug development and drug safety surveillance1-4.
The long-proclaimed aim of medicine to ’tailor the treatment to the patient‘ seems to be progressing most in cancer therapeutics. Small molecules and antibodies directed at specific molecular targets have shown striking or at least potentially useful efficacy with Tamoxifen for estrogen-receptor-positive breast cancers, Herceptin for Her2/neu over-expressing breast cancers, Gleevec for chronic myelogenous leukemia with bcl-abl t9;22 translocation (“Philadelphia chromosome”) and for gastrointestinal stromal tumors, Avastin for some patients with colorectal cancer, and EGF receptor antagonists for a small percentage of lung cancer patients who have certain kinds of mutations of the EGF receptor. Many others are in the pipeline. However, there is recognition of emergence of resistance to Gleevec, and there are criticisms5 that some of the new anti-cancer drugs have only a modest benefit at a very high cost. Universally, reviews of drug development lament the high fatality rate of promising drugs already in clinical development, partly for lack of efficacy, but mostly for unrecognized toxicity6.
Pharmaceutical agents are some of the most potent means of perturbation of cells or whole organisms, making multi-level systems biology studies of the susceptibility to disease and the responses to therapy highly desirable. Such systems studies can assay genome variation, gene methylation, gene expression (mRNA transcripts), protein expression and modifications, and then metabolite patterns7,8. These levels of investigation are certain to be complementary, as it is already well known that mRNA and protein expression changes are not highly correlated, due to the different half-lives and the very many post-translational modifications of proteins that affect degradation, elimination, and function.
The special role of proteomics
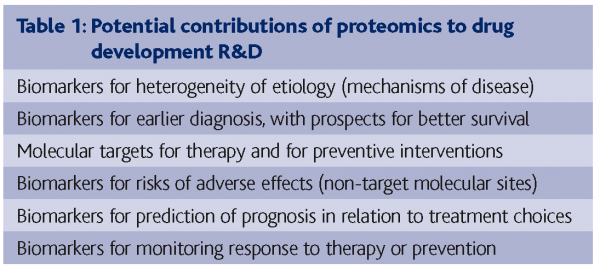
Proteomics of tissue specimens and of plasma specimens can address several key needs in drug development. Proteomics using mass spectrometry can be performed directly on tissue slices9, after laser capture micro-dissection to enrich for cell type, or on tumor lysates, to add molecular characteristics to the histological and immunohistochemical features that pathologists use to evaluate surgical specimens. Tissue proteomics will be complementary to mRNA gene expression patterns. Studies of target tissues for therapy and of other tissues can proceed with fewer complexities than studies of plasma, at least in pre-clinical, animal-based studies. For clinical studies, it is much harder to obtain liver, kidney, brain, or other target organ tissues, and impossible in the screening mode. For clinical studies, plasma and serum are the most accessible sources of new specimens and the most frequent material in specimen archives from clinical trials and epidemiological studies. Proteins in the circulation are a dynamic reflection of organ functions throughout the body. It is feasible to link these specimens to some kinds of clinical data, depending on the nature of the prospective study and the care with which IRB review and approval and informed consent from patients or study participants are obtained10.
Advances needed for plasma proteomics
Investigators need to be able to draw upon well-developed databases identifying the plasma and serum proteome in healthy persons, and then in specific diseases. The process of discovering candidate disease biomarkers has proved arduous, whether for individual proteins, panels of proteins, or patterns from large numbers of proteins, using various experimental and analytical methods11-14. Validation that proposed tests have high sensitivity (few false negatives), high specificity (few false positives and good differentiation from other similar kinds of disease), and high positive predictive value (credibility for clinical use) has been rare. In fact, the commonly used single-protein tests for prostate-specific antigen (PSA) for prostate cancer and CA-125 for ovarian cancer have such limited sensitivity and specificity and poor predictive value that all serious evidence-based reviews have recommended against their use for general population screening (see U.S. Preventive Services Task Force101. Long lists of proteins, like long lists of expressed genes, are hard to utilise unless translated into a comprehensible short list of the most differentiating proteins or expressed genes, or at least into a useful algorithmically defined ’pattern‘ and tied to functional pathways15. The Holy Grail for proteomics is the combination of high sensitivity (to detect low abundance proteins of tissue origin), high resolution (to separate the numerous proteins in a given specimen fraction), and high throughput (to make it feasible to analyse hundreds or thousands of specimens at reasonable cost in reasonable time).
The HUPO Plasma Proteome Project (Pilot Phase)
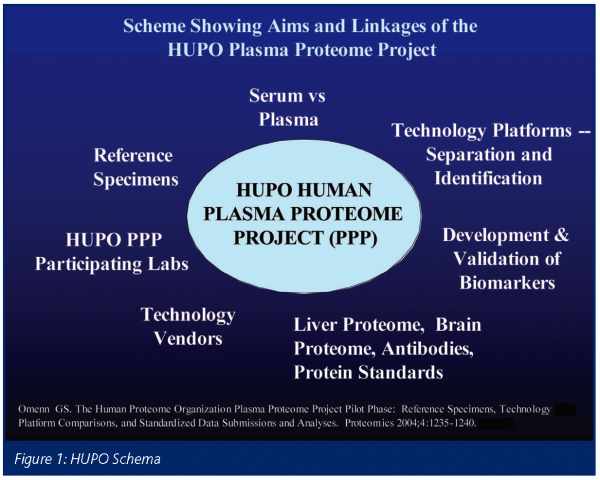
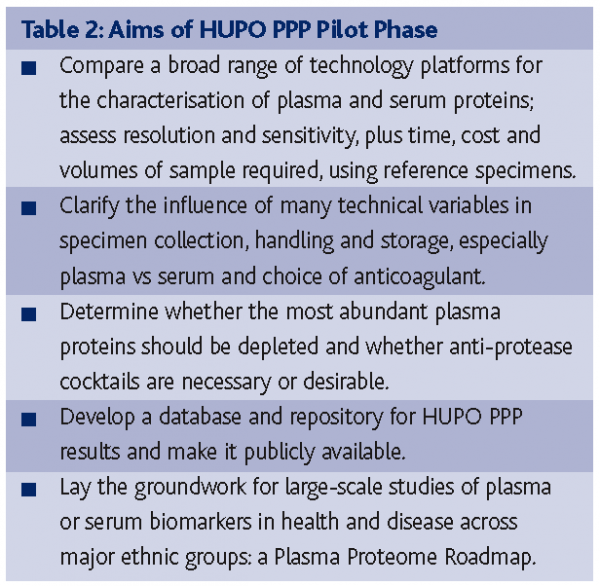
The Human Proteome Organization, launched in 200116, generated several major initiatives. As shown in Figure 1, HUPO has human plasma, brain and liver proteome projects plus initiatives for large-scale antibody production and for protein standards/bioinformatics. These are major global collaborations. The Plasma Proteome Project (PPP) has three long-term scientific goals: (1) a comprehensive analysis of plasma and serum proteins in humans; (2) identification of biological sources of variation within individuals over time, laying a foundation for discovery and validation of biomarkers in relation to physiological, pathological, and pharmacological responses; and (3) determination of the extent of variation across populations and within population, crucial for stratifying patients for treatment trials and for identifying co-variables and confounders for epidemiological studies and preventive trials17. The aims of the PPP Pilot Phase are shown in Table 2.
After intensive planning meetings, a kickoff Workshop was convened in Bethesda in July 2003. Altogether, 55 experienced laboratories in 14 countries requested one or more HUPO PPP reference specimens of plasma and serum and applied their own best technologies to the fractionation, analysis, and interpretation of the findings. For those using mass spectrometry to identify proteins, data were submitted to the PPP core bioinformatics unit at the University of Michigan for quality assurance, removal of duplicate protein IDs, and development of the Project-wide database. The results were subjected to cross-laboratory analyses at a Jamboree Workshop in June 2004 and multiple additional analyses in the subsequent months. A special issue of Proteomics titled “Exploring the Human Plasma Proteome” has been published in August 2005 with 28 articles from and about the PPP. About half of these articles document the collaborative experimental analyses, development of the database, and annotation of the database; the other half represent lab-specific studies that extend the findings in the collaborative study along lines identified by the investigators and the HUPO PPP steering committee, with seed grant funding to assist the investigators. The main findings of the issue are highlighted in the Overview paper from all the primary investigators18. The data are publicly available now at three websites: European Bioinformatics Institute102, Institute for Systems Biology103 and University of Michigan104.
In brief, 15,519 non-duplicated proteins were reported from the various analyses of 18 laboratories utilising tandem MS as the core technology; an “integration algorithm” developed for the PPP19 reduced this number to 9504 after choosing a single protein entry to represent however many proteins in the IPI v2.21 (July 2003) database20 matched perfectly to the relevant peptides. These figures were reduced to 5102 (unintegrated) and 3020 (integrated) when we applied the requirement that the protein match be based upon at least two peptides. We call this list the PPP Core Dataset, documented in accord with the guidelines proposed by Carr et al21. A more stringent set could require at least three peptides (1274 proteins, integrated). Interested users of the database can create many alternative lists by choosing different criteria for inclusion and exclusion, both from the MS search engine algorithm filters22 and from many kinds of annotations (proteins with transmembrane domains, proteins with signal sequences suggesting secretory origin, proteins with particular cellular localization or molecular process or biological function according to Gene Ontology, for examples)18,23.
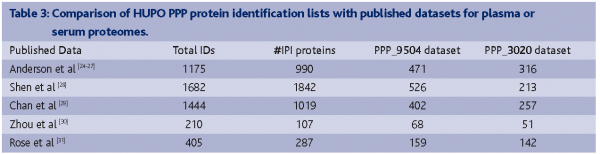
Certain HUPO analysts requested the entire collection of raw spectra and peaklists, not just peptide sequences, to perform independent analyses of the identifiable proteins. Given the desired, but very large, heterogeneity of approaches employed by the various laboratories, these independent analyses will provide more consistent lists without the major variables arising from individual investigators’ decisions to use different fractionation methods, different instruments, different search algorithms and filters for peptide identification, and different databases for protein assignment; numbers of proteins in the same range as our Core Data have been generated by these independent analyses18. Other substantial lists of proteins have been reported by individual laboratories not participating or not submitting data from the PPP collaboration, as summarized in Table 3. It is clear from these publications24-31 and our own collaboration that the various protocols and instruments give complementary results with only partial overlap. Some or many of these protein identifications may prove to be false-positives, or may be close to the limits of detection with the particular methods used, making it very difficult to replicate findings even with the same specimen or fraction of a specimen in the same lab. For example, 7-10 replicate MUDPIT MS/MS runs were needed to approach complete identification of peptides and proteins in work from the Yates and Schnitzer labs with endothelial cell preparations32.
Other HUPO PPP laboratories utilised protein microarrays or quantitative immunoassays, which enabled us to relate detectability of proteins to measured concentrations, with a high correlation between number of peptides identified and protein concentration (abundance)33. Ten laboratories utilised direct MS/SELDI analyses, with results reported34. Overall, plasma gave more reliable and reproducible results than serum; we recommend EDTA (or citrate) as anti-coagulant, rather than heparin18.
Potential next phases for the HUPO Plasma Proteome Project
Workshops at the 4th HUPO World Congress on Proteomics in Munich 27-28 August, 2005, will review progress and plans for each of the HUPO initiatives. There are many opportunities for the HUPO Plasma Proteome Project going forward.
First, the Proteomics August 2005 papers reveal several open questions which require more focused studies: (a) to generate guidelines and standardised operating procedures for specimen collection, handling, archiving and post-archive processing; (b) to optimise specific immuno-affinity depletion of abundant proteins with minimal non-target losses; (c) to combine separation platforms and MS capabilities with an aim to expand the portion of the plasma proteome that can be profiled with confidence, yet with higher throughput; (d) to achieve quantitative comparisons across specimens, not just identifications; (e) to achieve higher concordance for detectability and for concentrations in repeat analyses of the same specimen with the same methods; and (f) to overcome the extremely low overlap between protein identification datasets within a large collaboration of this type and, of course, across the literature, especially addressing the discrepancies due to post-MS/MS spectral analysis and peptide and protein database matching.
Other challenges are not specific to the plasma proteome. These will be evaluated together with other HUPO Initiatives: (a) to overcome the limitations of present sequence databases, which are incomplete, redundant, and constantly being updated with corrections and new splice variants and SNPs; (b) to improve the true-positive to false-positive ratio, which requires explicit optimisation; (c) to prepare reference specimen materials with specific objectives and user communities in mind; (d) to pursue independent corroboration of initial findings; and (e) to organise strategies to validate proteomic discoveries and lead to microarray analyses with well-characterized antibodies, so that large numbers of specimens from clinical trials and epidemiological studies can be assayed.
Second, the HUPO PPP could play a leading role in the continuing development and analysis of datasets arising from all quarters, in collaboration with the HUPO Protein Standards Initiative led by EBI and other bioinformaticians. We will start with cross-initiatives analyses of Human Liver Proteome and Human Brain Proteome datasets with the PPP datasets, explicitly including experimental analyses of plasma samples from the same people and animals whose liver and brain specimens are studied.
Third, there is an opportunity for HUPO to facilitate and possibly organise, major disease-related studies of candidate biomarkers for earlier diagnosis, better stratification of newly diagnosed patients, appropriate pathways-based monitoring of targeted therapies, and design of preventive interventions. For the overriding strategic question of gaining much higher throughput, at least four options have emerged in preliminary discussions: (a) LC-MS with highly accurate mass and elution time parameters for peptide identification35; (b) high accuracy LC-MS/MS/MS for peptide identifications, e.g. MS3, comprising MS/FTICR/MS36; (c) protein affinity micro-arrays37; and (d) isotope coded peptide standards for quantitative protein identification38.
Conclusions
Proteomics methods will enable profiling of tissues and plasma to generate useful panels or arrays of biomarkers for mechanistic insights, earlier diagnosis, treatment monitoring, and better prognosis. As organ-specific and agent class-specific ’signatures‘ of proteomics profiles emerge, it may be feasible to recognize risks of adverse effects much earlier in the drug development process. The Pilot Phase of the HUPO Plasma Proteome Project demonstrates that major advances toward the Holy Grail of simultaneous high resolution, high sensitivity and high throughput will be needed in order to characterise large numbers of specimens. Finally, much better replication of findings with a single specimen will be needed in order to reliably compare the proteome of two different specimens.
Acknowledgements
I thank the many investigators who contributed so much to the experimental datasets and analyses of the HUPO Plasma Proteome Project, as well as the corporate and federal (trans-NIH grant supplement CA-84942) sponsors18.
Web Sites
101. U.S. Preventive Services Task Force, www.ahrq.gov/clinic/uspstfix.htm
102. EBI, www.ebi.ac.uk/pride/hupo/ppp
103. ISB, www.systemsbiology.org/resources; www.peptideatlas.org/repository
104. UM, www.bioinformatics.med.umich.edu/ppp




References
- Stoughton RB, Friend SH. How molecular profiling could revolutionize drug discovery. Nature Review Drug Discovery 2005;4:345-350.
- Liu ET. Expression genomics and drug development: towards predictive pharmacology. Briefings Func Genomics Proteomics 2005; 3: 303-321.
- Mann M, Aebersold R. Mass spectrometry-based proteomics. Nature 2003; 422: 198-207.
- Hanash S. Disease proteomics. Nature 2003; 422:226-232.
- Avorn J. Sending pharma better signals. Science 2005;309:669.
- Ryan TP, Watson DE, Berridge BR. Toxicology biomarkers in drug development: delivering on the genomic promise. Pharmaceutical Discovery 2004; 4: 22-28.
- Hood L, Heath JR, Phelps ME, Lin B. Systems biology and new technologies enable predictive and preventative medicine. Science 2004; 306: 640-643.
- Robertson DG, Reilly MD. Metabonomics: evaluating drug safety. Euro Pharm Rev 2005; 10:28-34.
- Caldwell RL, Caprioli RM. Tissue profiling by mass spectrometry: a review of methodology and applications. Mol Cell Proteomics 2005;4:394-401.
- Nestler G, Steinert R, Lippert H, Reymond MA. Using human samples in proteomics-based drug development: bioethical aspects. Expert Rev Proteomics 2004; 1: 77-86.
- Jain KK. Role of oncoproteomics in the personalized management of cancer. Expert Rev Proteomics 2004; 1:49-55.
- Petricoin EF III, Ardekani AM, Hitt BA, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet 2002; 359: 572-577.
- Bouwman K, Qiu J, Zhou H, et al. Microarrays of tumor cell derived proteins uncover a distinct pattern of prostate cancer serum immunoreactivity. Proteomics 2003; 3: 2200-2207.
- Qiu J, Madoz-Gurpide J, Misek DE, et al. Development of natural protein microarrays for diagnosing cancer based on an antibody response to tumor antigens. J Proteome Research 2004; 3: 261-267.
- Marko-Varga G. Pathway proteomics: global and focused approaches. Amer J Pharmacogenomics 2005;5:113-122.
- Hanash SM, Celis JE. The Human Proteome Organization: a mission to advance proteome knowledge. Mol. Cell Proteomics. 2002;1:413-414.
- Omenn GS. The Human Proteome Organization Plasma Proteome Project Pilot Phase: Reference specimens, technology platform comparisons, and standardized data submissions and analyses. Proteomics 2004;4:1235-1240.
- Omenn GS, States DJ, Adamski M, et al. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics 2005;13:5 in press.
- Adamski M, Blackwell T, Menon R, et al. Data management and preliminary data analysis in the pilot phase of the HUPO Plasma Proteome Project. Proteomics. 2005;13:5 in press.
- Kersey, P.J., Duarte, J., Williams, A., Karavidopoulou, Y., Birney, E., & Apweiler, R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985-1988. http://www.ebi.ac.uk/IPI/IPIhelp.html.
- Carr S, Aebersold R, Baldwin M. The need for guidelines in publication of peptide and protein identification data. Mol Cell Proteomics. 2004;3:351-353.
- Kapp EA, Schutz F, Connolly LM, et al. An evaluation, comparison and accurate benchmarking of several publicly-available MS/MS search algorithms: sensitivity and specificity analysis. Proteomics. 2005;13:5 in press.
- Ping P, Vondriska TM, Creighton CJ, et al. A functional annotation of subproteomes in human plasma. Proteomics 2005;13:5 in press
- Anderson NL, Polanski M, Pieper R, et al. The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol Cell Proteomics. 2004;3:311-316.
- Pieper R, Gatlin CL, Makusky AJ, et al. The human serum proteome: display of nearly 3700 chromatographically separated protein spots on two-dimensional electrophoresis gels and identification of 325 distinct proteins. Proteomics. 2003;3:1345-1364.
- Adkins JN, Varnum SM, Auberry KJ, et al. Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol. Cell. Proteomics. 2002;1:947-952.
- Tirumalai RS, Chan KC, Prieto DA, Issaq H J, Conrads TP, Veenstra TD. Characterization of the low molecular weight human serum proteome.Mol Cell Proteomics. 2003;2:1096 –1103.
- Shen Y, Jacobs JM, Camp DG II, et al. Ultra-high-efficiency strong cation exchange LC/RPLC/MS/MS for high dynamic range characterization of the human plasma proteome. Anal. Chem. 2004;76:1134-1144.
- Chan KC, Lucas DA, Hise D, et al. Analysis of the Human Serum Proteome. Clinical Proteomics. 2004;1:101-226.
- Zhou M, Lucas DA, Chan KC, et al. An investigation in the human serum interactome. Electrophoresis. 2004;25:1289-1298
- Rose K, Bougueleret L, Baussant T, et al., Industrial-scale proteomics: from liters of plasma to chemically synthesized proteins. Proteomics. 2004;4:2125-2150.
- Durr E, Yu J, Krasinska KM, et al. Direct proteomic mapping of the lung microvascular endothelial cell surface in vivo and in vitro. Nature Biotechnol 2004; 22:985-992.
- Haab BB, Geierstanger BH, Michailidis G, et al. Immunoassay and antibody microarray analysis of the HUPO PPP reference specimens: systematic variation between sample types and calibration of mass spectrometry data. Proteomics. 2005;13:5 in press.
- Rai AJ, Stemmer PM, Zhang Z, et al. Analysis of HUPO PPP reference specimens using SELDI-TOF mass spectrometry: multi-institution correlation of spectra and identification of biomarkers. Proteomics 2005;5 (in press
- Adkins JN, Monroe ME, Auberry KJ, et al. A proteomic study of HUPO’s Plasma Proteome Project pilot samples using an accurate mass and time tag strategy. Proteomics 2005; 5 (in press).
- Olsen JV, Mann M. Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc. Natl. Acad. Sci. U.S. 2004;101:13417-13422.
- Humphery-Smith I, A human proteome project with a beginning and an end. Proteomics 2004;4:2519-2521.
- Aebersold R. Constellations in a cellular universe. Nature 2003;422:115-116.




