Initiatives from Spain
Posted: 22 August 2005 | | No comments yet
The availability of the complete sequence of some model organism genomes, including the human genome, offers new opportunities for biological research. The goal is to establish technology to identify all the proteins involved in a particular biological process and the interactions between them.
The availability of the complete sequence of some model organism genomes, including the human genome, offers new opportunities for biological research. The goal is to establish technology to identify all the proteins involved in a particular biological process and the interactions between them.
The availability of the complete sequence of some model organism genomes, including the human genome, offers new opportunities for biological research. The goal is to establish technology to identify all the proteins involved in a particular biological process and the interactions between them.
This is a very important technology, as diseases express themselves at protein level rather than gene level. By comparing proteins present in diseased and healthy tissue (or body fluid samples), it is possible to identify the specific ’proteomic signature’ of a disease, which may represent potential therapeutic targets or molecular markers for the progress of a disease or the activity of a drug. Proteomic research covers the separation, identification and characterisation of the proteins present in a biological sample and the interactions between the proteins of a cell, identifying all the points at which a specific metabolic process can be modified and those at which potential new therapies can be directed.
A focused facility
CIC bioGUNE is a research centre that concentrates on conducting basic, cooperative, multidisciplinary research in strategic areas of biosciences. From its very beginning CIC bioGUNE has focused its interest in proteomics and is now involved in coordinating two major proteomic initiatives: it is the first node of HUPO (Human Proteome Organisation) in Spain, named EHUPO and it is also the international coordinator of the Spanish National Network for proteomic services: PROTEORED, financed by the Spanish Genome Foundation and coordinated by the Centro Nacional de Biotecnología (CNB-CSIC) in Madrid. The Proteomics Core Facility at CIC bioGUNE includes cutting edge technology and senior researchers for the more advanced proteomics research.
The CIC bioGUNE proteomics core facility is focused on the study of proteins and proteomes by mass spectrometry. The main aim of this unit is to give technical support in this field and allow the different research groups from CIC bioGUNE and the scientific community in general to carry out proteomics studies. Moreover, it also hosts different research projects. Among them and within the frame of the HUPO project, CIC bioGUNE is involved in the study of the human serum and liver proteome.
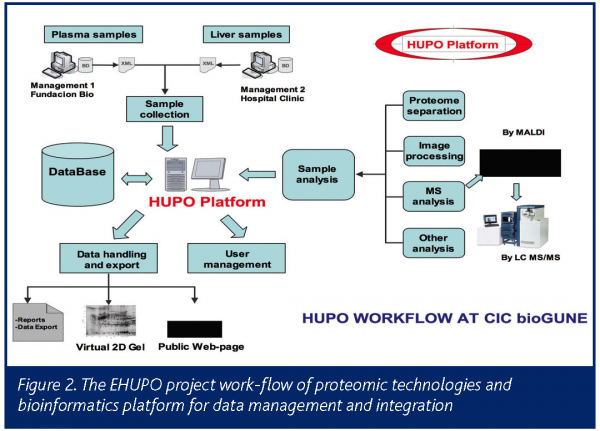
Through EHUPO, CIC bioGUNE coordinates a number of research groups at the University, research centres, hospitals and private companies for the identification and characterisation of new proteins in both plasma and liver, which could be used as diagnostic markers for different diseases.
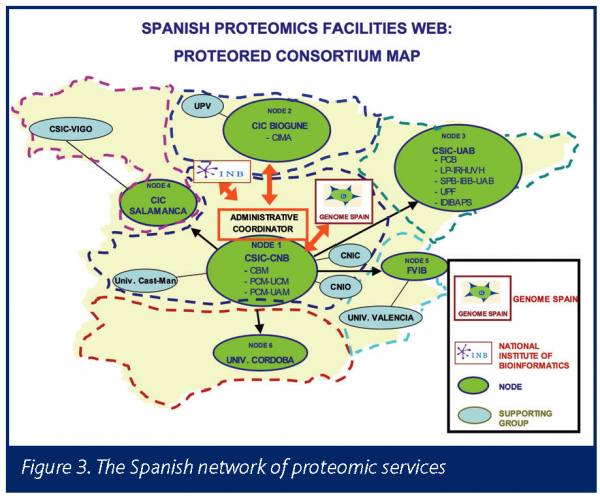
The second proteomics initiative at CIC bioGUNE is the PROTEORED project, coordinated by Dr. Juan Pablo Albar at the CNB-CSIC in Madrid. PROTEORED is composed of a strong consortium with a nodal structure of six nodes, but integrating and supporting more than 20 proteomic facilities providing services across Spain. This project is aimed at the coordination, integration and development of proteomic services in different Spanish regions.
HUPO Spanish Initiative
The Spanish HUPO project – named EHUPO – is coordinated by CIC bioGUNE. This initiative started with the creation of a powerful scientific/technological partnership formed by 15 partners. These partners are from the academic and health sectors, including some private companies from the Spanish biosciences sector, though this partnership is opened to future incorporations of other research groups in proteomics. The funding of EUR2.8 Million for three years has been granted by: BBVA Foundation, Basque Government, Bizkaia County Council and the Spanish General Administration.
The Spanish consortium has decided to actively participate in the subprojects of plasma (HPPP) and liver (HLPP). The main objective of EHUPO project is the identification and characterisation of new plasma and liver proteins for the discovery of new diagnosis markers in different diseases, with two specific aims:
- Identification of new plasma proteins associated to albumin and other transporting proteins
- Identification and characterisation of human proteins related to clinically described hepatic pathologies
The main activities of EHUPO partners planned for the next three years include:
- Standardisation of the criteria and protocols for the extraction of proteins of the selected biological systems
- Collection of liver and plasma samples both in healthy and in pathological conditions
- Analysis of plasma and liver proteins
- Definition of expression patterns associated to the different pathologies
- Identification of the sub-cellular and tissular location of the identified proteins
- Development of a bioinformatics platform for the integration and management of the generated data
CIC bioGUNE is coordinating the EHUPO project in close contact with the general coordinator of the international HUPO project in Montreal and participates in the annual HUPO meetings, presenting the results of the Spanish project. The identified proteins will be available on the CIC bioGUNE Web site.
PROTEORED initiative
The main objective of PROTEORED is to coordinate and integrate the activities of the Spanish proteomic facilities and services in a common network with a nodal structure, so they will support the development of proteomic research in Spain. PROTEORED will increase the specialisation and competitiveness of the proteomic facilities, considering the type of technologies/equipment offered, the type of customers, the geographical situation and their expertise, trying to provide the best service to the highest number of research groups.
With the proposed objectives in mind, seven working areas were constituted, composed of area experts from the six nodes, with the aim of achieving all PROTEORED goals. In addition the working areas support the proteomic facilities and services of the six nodes, with the idea of extending their supporting to the 20 proteomic facilities. The six defined working areas for supporting the proteomic services are:
- Technological development and standardisation of protocols
- Sample collection and handling
- Bioinformatics supporting area
- Functional organisation of proteomic services and coordination of price scales
- Training, education and diffusion
- Internationalisation and coordination with other proteomic consortia or international platforms
PROTEORED also has the objective of testing technological developments for providing new proteomic methodologies and equipment to the Spanish proteomic facilities. It will also establish open channels with the customers of these proteomic services to know their technological needs, accuracy of the data, quality requirements, price scales and services needed for the future.
PROTEORED network will offer major services necessary in all the stages of the protein analysis process: proteins fractionation, separation and purification of peptides and proteins, proteins identification, quantitative determination of peptides and proteins, protein sequencing, complex analysis of proteins, peptide mass fingerprinting, analysis of mass spectrum, analysis of images and peptide synthesis.
The funding provided by GENOMA ESPAÑA Foundation for the first year is EUR1 Million, with five years funding up to EUR6.5 Million and, as established by GENOMA ESPAÑA guidelines, the main part of the funding will be dedicated to personnel, reaching 82 per cent of the funding for contracting 22 new technicians. Since the very beginning, PROTEORED was conceived to be sustainable after five years of funding by GENOMA ESPAÑA Foundation.
Proteomic services and facilities at CIC bioGUNE
The Proteomics Core Facility at CIC bioGUNE harbours an entire two dimensional gel electrophoresis system for differential expression analysis in the same gel and also a two dimensional chromatographic system for automatic protein separation and further analysis. Once proteins are separated they can be identified and characterised by the in-house Proteomics Core Facility. This lab is focused on mass spectrometry analysis and is equipped with a medium/high throughput protein identification platform consisting in a robotic workstation for protein digestion and further sample preparation and a MALDI TOF/TOF mass spectrometer for protein identification. There is also a nano UPLC system coupled to a Q-TOF type of instrument for in-depth protein characterisation (e.g. post translational-modification analysis as phosphorylation, etc.) or complex peptide mixture analysis. The Proteomics Core Facility has a Web page with the following features: list of services, interactive electronic form for service request, sample requirements and delivery instructions and user tips such as staining and storage and spot excision.
The Proteomics Core Facility is actively involved in the HUPO project and maintains a tight collaboration with the Metabolomics Unit at CIC bioGUNE and with OWL Genomics (www.owlgenomics.com) to set up technologies intended to identify all the metabolites involved in a specific metabolic route by mass spectrometry (PROMETA Core Facility). Metabolomics is a highly important emerging discipline, as numerous diseases express themselves at the level of specific metabolites. By comparing the metabolites present in normal and diseased tissues (or body fluid samples), metabolic fingerprint of a disease can be established and altered metabolic pathways can be identified. Thus, potential new therapies can be designed.
Research activities
Our laboratory has played a key role in demonstrating that a chronic deficiency in hepatic S-adenosylmethionine (SAM) favours the development of non-alcoholic steatohepatitis (NASH) and its progression to cirrhosis and hepatocarcinoma (HCC). We generated a knockout mouse deficient in hepatic SAM (MAT1A-KO) that allowed us to explore the molecular mechanisms that lead to the generation of NASH and HCC (Lu et al., 2001). We obtained a genomic signature of NASH (85 genes) through the comparison of genome wide, gene expression data of liver samples from patients with early stage NASH and knockout mice at various times during the development of the disease. One of these genes encodes for Sp1, suggesting that this transcription factor is involved in the mechanisms that lead to the development of NASH. The role of Sp1 in the regulation of the expression of these genes was confirmed by chromatin immunoprecipitation (ChIP) assay and it was also determined that Sp1 is hyperphosphorylated in the liver of MAT1A-KO mice prior to the clinical manifestation of NASH. Recent data points to Sp1 as a key mediator of the cross-talk between specific signaling pathways through the regulation of the expression of gene targets of these pathways, being its transcriptional activity dependant on the phosphorylation status. These and other results support the hypothesis that SAM is a regulator of the phosphorylation/dephosphorylation of proteins that regulate cell metabolism, gene expression and cell proliferation and that alterations in this function, as a consequence of a chronic deficiency in hepatic SAM, facilitate the development of NASH and its progression to HCC.
Sp1 interacts with other proteins and transcription factors in the chromatin. It has been postulated that the cellular and temporal specificity of the regulation of gene expression in response to the phosphorylation of Sp1 is determined by this set of interactions. Consequently, we are working on the identification, through the employment of ChIP assays combined with proteomic techniques (2D gels and mass spectrometry), of the network of proteins associated to Sp1 in the chromatin, the ’Sp1-proteome‘ and its differential expression in liver samples of MAT1A-KO mice at different ages and stages of the NASH.
Our group has also carried out proteomic studies on livers from MAT1A-KO mice, finding 117 proteins that were differentially expressed when compared to livers from wild-type mice (Santamaria et al., 2003). Given the importance of SAM in phosphorylation, we decided to further study the MAT1A-KO mouse liver proteome, focusing on the differential expression of phosphoproteins, to try to identify the signaling pathways that are altered. Once we identify the pathways, we will try to confirm the results in liver samples from patients with NASH. To this end, we are using a combination of two techniques: phosphatase treatment and difference gel electrophoresis (DIGE). DIGE enables multiplexing up to three different protein mixtures, differentially labeled with three fluorophores. One of these protein mixtures is an internal standard, made by pooling all samples and it is used to match protein patterns across gels avoiding the problem of inter-gel variation. Therefore, DIGE allows for accurate quantification of statistically significant differences between samples. Proteins of interest are identified using mass spectrometry.




Bibliography
Lu SC, Alvarez L, Huang ZZ, Chen L, An W, Corrales FJ, Avila MA, Kanel G and Mato JM (2001) Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci U S A 98:5560-5565.
Santamaria E, Avila MA, Latasa MU, Rubio A, Martin-Duce A, Lu SC, Mato JM and Corrales FJ (2003) Functional proteomics of nonalcoholic steatohepatitis: mitochondrial proteins as targets of S-adenosylmethionine. Proc Natl Acad Sci U S A 100:3065-3070.
Fernández-Irigoyen J, Santamaría E, Sesma L, Muñoz J, Riezu I, Caballería J, Lu SC, Prieto J, Mato JM, Avila MA, Corrales FJ (2005) Oxidation of specific methionine and tryptophan residues of apolipoprotein A-I in hepatocarcinogenesis. Protemics, in press




