RNAi-based therapies for the treatment of HIV
Posted: 24 June 2010 | | No comments yet
Since the discovery of RNA interference (RNAi) in 1998 and the demonstration of RNAi in mammalian cells in 2001, research into the mechanisms and applications of this pathway has moved swiftly. RNAi is capable of mediating potent and specific silencing of genes and has therefore shown promise in the development of alternative anti-viral therapies with the potential to avoid disadvantages associated with conventional drug regimens. A number of synthetic and expressed constructs have been investigated against HIV with varying success. Despite rapid progress, important hurdles need to be surmounted before a safe, effective and widely applicable therapy can be implemented clinically. Here, we review different RNAi-based strategies against HIV and highlight future developments necessary for the realisation of an effective anti-HIV therapy…
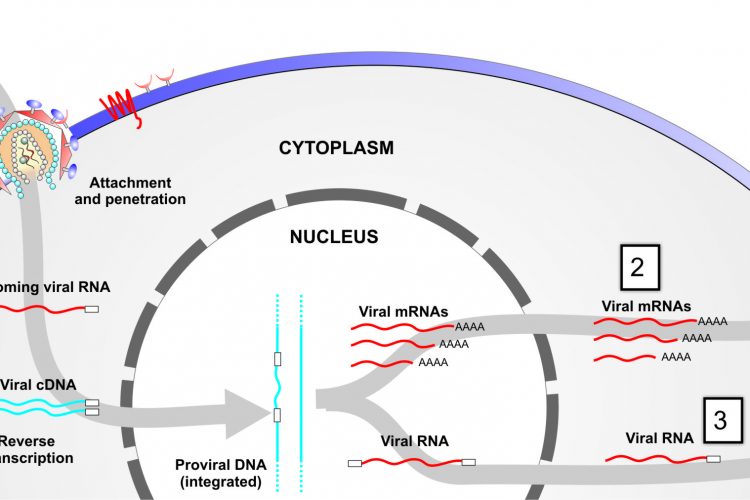

Figure 1 Targeting viral-associated RNAs at different stages of infection. Step 1, targeting of incoming RNA; step 2, targeting of viral mRNAs following provial integration; and step 3, targeting viral outgoing pregenomic template DNA.
Since the discovery of RNA interference (RNAi) in 19981 and the demonstration of RNAi in mammalian cells in 20012, research into the mechanisms and applications of this pathway has moved swiftly. RNAi is capable of mediating potent and specific silencing of genes and has therefore shown promise in the development of alternative anti-viral therapies with the potential to avoid disadvantages associated with conventional drug regimens. A number of synthetic and expressed constructs have been investigated against HIV with varying success. Despite rapid progress, important hurdles need to be surmounted before a safe, effective and widely applicable therapy can be implemented clinically. Here, we review different RNAi-based strategies against HIV and highlight future developments necessary for the realisation of an effective anti-HIV therapy.
Human immunodeficiency virus (HIV) infection is a serious epidemic worldwide, but is particularly rife in sub-Saharan Africa, which harbours approximately 65 per cent of all HIV infected individuals and mortality due to AIDS related deaths was approximately 1.4 million in 2008 alone3. The success of antiviral cocktails used in highly-active antiretroviral therapy (HAART) is unmatched in modern medicine; these drugs have been developed with unprecedented speed and have dramatically reduced the morbidity and mortality of HIV-related illness. HAART has been credited for demystifying HIV as an ‘untreatable virus’ and has relegated HIV/AIDS to a manageable chronic disease. Nevertheless, current drug regimens have many shortcomings. Several antiviral drugs have serious toxic side effects, and patients over the long-term are likely to require newer and more expensive cocktails to circumvent viral drug resistance. Furthermore, in resource-poor settings, patient compliance to therapy is often lacking and the inability to purge cellular ‘reservoirs’ of latent infection means that patients will need to be treated for their entire lives. The future of HIV therapy, therefore, rests on developing an entirely new therapeutic paradigm.
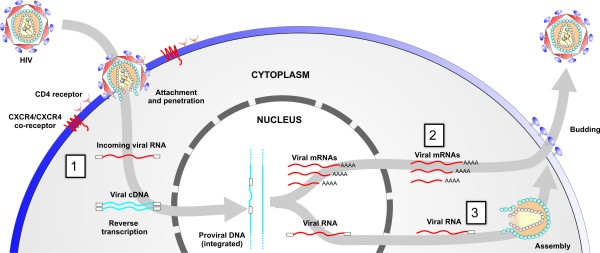

Figure 1 Targeting viral-associated RNAs at different stages of infection. Step 1, targeting of incoming RNA; step 2, targeting of viral mRNAs following provial integration; and step 3, targeting viral outgoing pregenomic template DNA.
RNA interference (RNAi)-based therapeutic approaches have emerged out of the shadow of previous nucleic-acid technologies, specifically antisense- and ribozyme-based inhibitors, which function by blocking viral transcription or translation through Watson-Crick comple – mentary interactions. While these earlier technologies were shown to be safe in vivo, a number of Phase I/II clinical trials have shown that their general efficacy as good antiretrovirals is inconsistent. RNAi therapies differ from other nucleic acid inhibitors in that they function through a natural cellular pathway whereby double-stranded RNA acts as a trigger for sequence-specific inhibition of gene expression. RNAi-based drugs block gene expression at the post-transcriptional level by engaging in an enzymatic cascade (involving Dicer and Argonaute proteins) which results in the targeted degradation of specific RNAs. Not surprisingly, from the onset, RNAi-based therapeutics performed better as gene knockdown agents compared to their ribozyme/antisense predecessors and shows the most promise for clinical development against HIV.
Where and what to target?
RNAi inhibitors can theoretically target several different stages of the HIV-1 lifecycle, from incoming viral RNA to de novo viral progeny (genomic) RNA and viral mRNAs (Figure 1 on page 33). The latter two are expressed following proviral integration in the host cell genome. Unfortunately, incoming viral RNA appears to be intransigent to RNAi-mediated attack, possibly because viral reverse-transcription is shielded by capsid and/or host proteins after fusion and entry. Blocking the virus prior to entry is an important objective as it ensures that the cell is completely protected from infection. Nevertheless, targeting multiple highly conserved regions within nascent viral RNAs prevents progeny virus formation, blocks early and late stage proteins and significantly diminishes further infection and replication.
Another viable approach is to target host genes known to be important for infection and propagation. Unlike viral genes, which readily mutate via the error-prone viral reversetranscriptase, host genes do not, hindering the development of inhibitor-resistant viral strains. Moreover, as highlighted already, factors associated with the viral life cycle prior to proviral integration represent particularly attractive targets. For example, the chemokine co-receptors CCR5 and CXCR4 are key components of viral entry. CCR5 in particular is known to be dispensable to humans, as healthy individuals with natural homozygous deletion variants of this gene exist in the general population. However, inhibiting CCR5 would only protect CD4+ T cells from R5-tropic virus. Since HIV often switches tropism during the course of infection, a more robust host factor is needed for widespread inhibition. New efforts aimed at elucidating novel host factors have emerged from genome-wide RNAi screens. We are still at an early stage with these new developments; currently little concordance exists between genes identified in several recent separate HIV screens4,5. RNAi screening still yields many false-positive hits, underscoring the need to functionally validate selected genes in the context of a physiologically relevant HIV infection. Once a specific host factor target is identified, any deleterious consequences associated with its inhibition must be clarified before further clinical progression is possible. Despite these concerns, our ability to identify important new targets for anti-HIV drug development has expanded significantly with these technological advances.
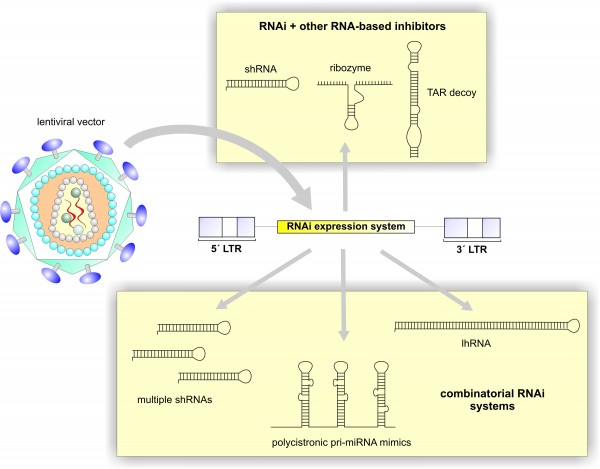

Different approaches for RNA-based therapies
The therapeutic application of RNAi for the treatment of HIV has followed two distinct strategies. RNAi can be elicited by using synthetic short interfering RNAs (siRNAs), which are 21 bp products of the RNAi pathway; or by introducing gene expression cassettes which transcribe siRNA precursors. SiRNA precursors are often introduced as short hairpin RNAs (shRNAs) or primary microRNA mimics (pri-miRNAs), which are expressed and processed by the RNAi machinery to generate active siRNAs in the cell. Each approach has its own benefits and shortcomings with respect to application (delivery), treatment duration and possible toxic side effects (see Table 1) as discussed below.
Gene-expressed RNAi inhibitors
The ability to express RNAi effectors from an expression cassette allows for the possibility of a more sustained, durable suppression of HIV through the continued renewal of RNAi precursors. This is important as it raises the prospect of introducing a single intervention therapy into patients. RNAi expression cassettes are also compatible with highly efficient viral vectors (such as lentiviral vectors), which may allow exploitation of the cell-specific targeting, integration and expression characteristics of these recombinant viruses. The focus in recent years has been to develop a combinatorial RNAi expression strategy whereby several different viral (or host factor) sequences are targeted simultaneously. This is achieved using either multiple shRNA-expressing units or single promoter cassettes expressing several RNAi precursors. Single promoter combinatorial RNAi systems have the advantage of not overwhelming the endogenous RNAi pathway, which can result in serious toxic side effects6 and examples include long hairpin RNAs (lhRNAs)7,8 and polycistronic miRNA mimics9 (Figure 2).
Another gene expression strategy aims to combine RNAi units with other technologies, such as ribozymes, TAR RNA decoys and/or dominant negative mutants of HIV Rev. The latter approach is being attempted in a phase I clinical trial by John Rossi and colleagues at the City of Hope Medical Center in Duarte, California. This trial involves a pilot study in AIDS lymphoma patients to assess the safety and therapeutic feasibility of using a self-inactivated lentiviral vector encoding three anti-HIV modalities, including an shRNA targeted to the tat/rev overlapping region, a ribozyme to CCR5 and a TAR RNA decoy10,11 (Figure 2). The procedure involves removal of bone marrow lymphocytes from patients and enrichment for CD34+ haematopoetic progenitor cells (HSCs). HSCs are transduced ex vivo with a lentivirus expressing the three transgenes before expansion and autologous reintroduction into patients who have undergone bone marrow ablation10 (Figure 3). While previous trials using ribozyme and antisense sequences suggest that this approach is likely to be well-tolerated in patients, it remains to be seen whether the transduced HSCs maintain their self-renewal properties in vivo and if long term viral suppression is possible. The potential therapeutic benefits of ex vivo gene therapy are great but one significant drawback is the cost and complication of such a medical intervention. No matter how successful, it is highly unlikely that such a therapy will be implemented in HIV-endemic areas where far simpler treatment methods are required.
Table 1 A general comparison of therapeutic RNAi strategies
| Synthetic RNAi Approaches | Gene-based RNAi Approaches | |
| Transient vs long-term inhibition | Transient effects, useful for periodic treatment. Multiple treatments are required. | Enduring effects, useful for long-term treatment. Potentially a once-off treatment. |
| Effect on natural RNAi pathway | Better control of dose | High expression levels can lead to saturation of the RNAi pathway and lethal toxicity. |
| Targeting host factors | High level expression is probably best avoided in targeting of host factors. | Regulated expression makes these effectors more appropriate for targeting of host factors. |
| Combinatorial RNAi | Several different siRNAs can be delivered simultaneously, but differential uptake could give inconsistent dosing of individual targets. | A single expressed construct can target several genes, with consistent dosing of individual targets as part of a combinatorial approach. |
| Biosyntheisis in vivo | Therapeutic silencing is dependent on a single RNAi processing mechanism which can limit targeting strategies. | A variety of different types of RNA-based effectors can be included in one expression cassette which can be useful for potent viral targeting (e.g. short hairpin & a ribozyme). |
| Chemical alterations | SiRNAs can be chemically modified, which might allow for enhanced therapeutic properties (e.g. delivery, efficacy, immunostimulation) | Processing of RNA-based effectors is dependent on intrinsic, natural properties of the effector sequences. |
| Delivery | SiRNAs must be complexed to nanoparticles or liposome non-viral vectors or conjugated to aptamer motifs | Expressed RNAi effectors require more concise delivery to specific cells for correct expression. Success is dependent on development of effective viral delivery vectors. |
Synthetic RNAi inhibitors
Synthetic siRNA do have some important advantages over expressed RNAi effectors. Importantly, siRNAs can be chemically modified to improve their efficacy in vivo. This includes increasing resistance to serum nucleases, enhanced strand selection by the RNAi machinery and preventing stimulation of the innate immune system. Unfortunately, the development of clinically viable synthetic siRNAs for use against HIV has been slow. There are fundamentally two reasons for this. Firstly, administration of siRNAs has been limited to diseases where there is ease of access or where surface application is feasible. Naked siRNA cannot efficiently be delivered to cells systemically in vivo. Secondly, until recently and with the exception of primate HIV models, there have not been any effective small animal models in which HIV replication and disease progression can be studied in vivo. The latter has been a serious impediment towards the clinical testing of systemic and/or targeted siRNA delivery strategies.
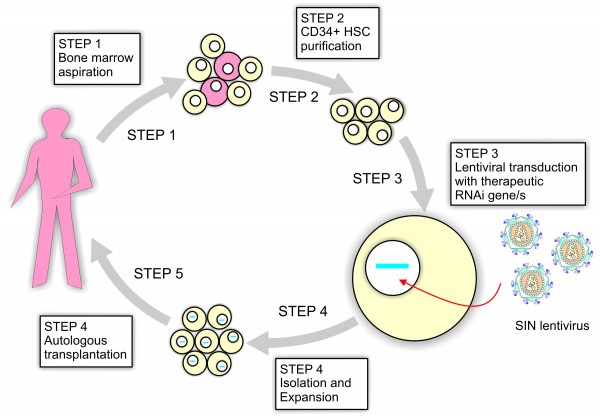

Figure 3 A method for ex vivo gene therapy. Transduced haematopoetic stem cells (HSCs) express anti-HIV therapeutic expression cassettes which are transplanted autologously into patients.
Recent advances in developing humanised immunodeficient mice have accelerated the development of anti-HIV siRNA therapeutics. A lot of recent attention has been given to non-obese diabetic (NOD) mice with severe combined immunodeficiency (SCID) and which have a complete null mutation in the common cytokine receptor γ chain that abrogates interleukin 2 binding (NOD/SCID/IL2rγ-/-). Another model involves transplantation of thymic and liver tissues under the kidney capsule of SCID mice (BLT mice). These mice are receptive to transplantation with human peripheral blood leukocytes (huPBLs) or human CD34+ HSCs (huHSC) which give rise to both myeloid and lymphoid cell lineages, including CD4+ T cells12. BLT mice additionally produce B cells that are capable of primary immune responses. Both mice models are capable of reconstituting a basic human immune system and can support active HIV-1 infection over several months, mimicking to some degree the natural progression of infection in humans (high plasma viraemia and gradual loss of CD4+ T cells over time).
Using these newer mice models, a lot of recent progress in the field of synthetic antiviral RNAi modalities and novel non-viral delivery tools comes from work by Premlata Shankar and colleagues. In a seminal study, a single-chain antibody specific for the T-cell specific CD7 receptor was conjugated to several anti-HIV siRNAs via an arginine tag13 (Figure 4 on page 36). Impressively, anti-vif and anti-tat siRNAantibody conjugates were introduced into NOD/SCID/IL2rγ-/- and SCID-hu mice and prevented HIV infection and CD4+ T cell depletion without any observed toxicities. However, since CD7 expression is only found on T cells, it is possible that other HIV-susceptible cells will not be targeted. To address this concern, the same group has recently developed a stabilised liposome nanoparticle consisting of neutral phospholipids and a singlechained antibody directed to the human lymphocyte function-associated antigen-1 (LFA-1), the predominant integrin found on all leukocytes (Figure 4 ). This LFA-1 targeted liposome nanoparticle was able to deliver anti-CCR5 siRNAs to T cells and macrophages in vivo whilst reducing viraemia and protecting CD4+ T cell loss14. Another interesting but far less developed technology involves the use of anti gp120 aptamer-siRNA chimeras15. Interestingly, the aptamers have the dual function of neutralising new infections and introducing anti-HIV siRNAs into infected CD4+ T cells (Figure 4). Despite the optimism, there are still hurdles to overcome. Little is known about the consequences of repeat administration of liposomes or nanoparticles. These complexes need to avoid detection by the adaptive immune system. Additionally, siRNAs that are introduced into cells via endosomal compartments must ensure that they do not elicit an innate immunostimulatory response. While some of these effects can be negated by chemical modification, clearly caution is needed during these exciting times for synthetic siRNA anti-HIV therapies.
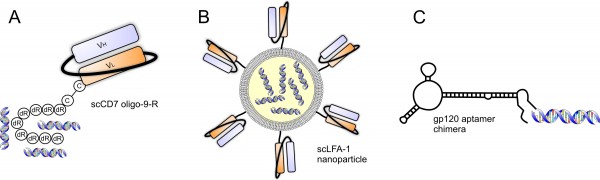

Figure 4 Synthetic siRNA-based delivery approaches. A) Single-chained antibodies to CD7 consisting of an argentine tag binds to negatively charged siRNAs. B) A stabilised nanoparticle consisting of a neutral phospholipids encapsulating siRNAs and targeted to the integrin LFA-1 via conjugated LFA-1 sing-chained antibodies. C) A gp120-targeted aptamer-siRNA chimera.
Conclusions
RNAi-based HIV therapeutics have progressed swiftly over the last eight years and already we await the outcome of an ex vivo gene therapy phase I clinical trial. Nevertheless, there are significant barriers that need to be overcome for progression towards the clinic. Technical hurdles include optimising dose control, limiting offtarget effects and improving in vivo delivery of RNAi effectors to HIV-susceptible cells. Moreover, careful consideration needs to be given to therapies that have widespread application, allowing future implementation in countries with the highest HIV/AIDS burden. In this regard, synthetic siRNA approaches appear to have the upper hand over ex vivo methods. Future improvements aimed at developing cell-targeted viral delivery approaches or safe mobilisation-competent viral vectors will ensure direct delivery of RNAi-based gene expression systems to patients – a technology which promises to revolutionise antiviral therapeutics. Rapid advances in the development of RNAi-based strategies against HIV could show the full potential of such a therapy sooner than expected.
References
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC: Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391(6669):806-811.
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T: Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411(6836):494-498.
- UNAIDS: AIDS Epidemic Update: November 2009. 2009:1-100.
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ: Identification of host proteins required for HIV infection through a functional genomic screen. Science 2008, 319(5865):921-926.
- Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE et al: Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 2008, 135(1):49-60.
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA: Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 2006, 441(7092):537-541.
- Saayman S, Barichievy S, Capovilla A, Morris KV, Arbuthnot P, Weinberg MS: The efficacy of generating three independent anti-HIV-1 siRNAs from a single U6 RNA Pol III-expressed long hairpin RNA. PLoS ONE 2008, 3(7):e2602.
- Saayman S, Arbuthnot P, Weinberg MS: Deriving four functional anti-HIV siRNAs from a single Pol III-generated transcript comprising two adjacent long hairpin RNA precursors. Nucleic Acids Res (In Press).
- Liu YP, Haasnoot J, ter Brake O, Berkhout B, Konstantinova P: Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res 2008, 36(9):2811-2824.
- Li M, Li H, Rossi JJ: RNAi in combination with a ribozyme and TAR decoy for treatment of HIV infection in hematopoietic cell gene therapy. Ann N Y Acad Sci 2006, 1082:172-179.
- Castanotto D, Rossi JJ: The promises and pitfalls of RNA-interference-based therapeutics. Nature 2009, 457(7228):426-433.
- Pearson T, Greiner DL, Shultz LD: Humanized SCID mouse models for biomedical research. Curr Top Microbiol Immunol 2008, 324:25-51.
- Kumar P, Ban H, Kim S, Wu H, Pearson T, Greiner DL, Laouar A, Yao J, Haridas V, Habiro K et al: T cellspecific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell 2008, 134:1-10.
- Kim SS, Peer D, Kumar P, Subramanya S, Wu H, Asthana D, Habiro K, Yang YG, Manjunath N, Shimaoka M et al: RNAi-mediated CCR5 silencing by LFA-1-targeted nanoparticles prevents HIV infection in BLT mice. Mol Ther 2010, 18(2):370-376.
- Zhou J, Rossi JJ: Aptamer–targeted cell-specific RNA interference. Silence 2010, 1(1):4.
About the Author
Marc S. Weinberg
Having completed a postdoctoral fellowship with leading RNA biologist Professor John Rossi at the City of Hope Medical Center in Los Angeles, Dr Marc Weinberg has, since 2005, led a group within the Antiviral Gene Therapy Research Unit (AGTRU) at the University of the Witwatersrand, South Africa. He and collaborators in California and Oxford are currently working on developing a range of molecular tools using RNAi directly to inhibit genetic elements that cause disease. At the AGTRU, his group’s specific emphasis is to target the Human Immunodeficiency Virus (HIV) and Hepatitis B Virus (HBV), both of which cause significant mortality globally – particularly in sub-Saharan Africa. Dr Weinberg and his team are tackling important challenges, which include designing genetic systems that effectively generate multiple RNAi-based drugs and solving the problem of RNAi delivery to cells which harbour virus.
Contact the Author
email: [email protected]





