Article 2: Reverse Transcription – a necessary evil
Posted: 20 March 2009 | Tania Nolan, Global Manager of Applications and Technical Support, Sigma-Aldrich and Stephen Bustin, Professor of Molecular Science, Centre for Academic Surgery, Institute of Cell and Molecular Science, Barts and the London School of Medicine and Dentistry | No comments yet
The fluorescence-based quantitative real-time polymerase chain reaction (qPCR)1-3, is the most widely used method to detect and measure minute amounts of DNA in a wide range of samples extracted from numerous sources. Since all currently available thermostable polymerases are DNA-dependent, RNA must be converted (“reverse transcribed”) into DNA prior to its amplification reaction. Both qPCR and reverse transcription (RT)-qPCR have revolutionised life sciences, agriculture, medical research and diagnostic and forensic applications4,5.
The fluorescence-based quantitative real-time polymerase chain reaction (qPCR)1-3, is the most widely used method to detect and measure minute amounts of DNA in a wide range of samples extracted from numerous sources. Since all currently available thermostable polymerases are DNA-dependent, RNA must be converted ("reverse transcribed") into DNA prior to its amplification reaction. Both qPCR and reverse transcription (RT)-qPCR have revolutionised life sciences, agriculture, medical research and diagnostic and forensic applications4,5.
The fluorescence-based quantitative real-time polymerase chain reaction (qPCR)1-3, is the most widely used method to detect and measure minute amounts of DNA in a wide range of samples extracted from numerous sources. Since all currently available thermostable polymerases are DNA-dependent, RNA must be converted (“reverse transcribed”) into DNA prior to its amplification reaction. Both qPCR and reverse transcription (RT)-qPCR have revolutionised life sciences, agriculture, medical research and diagnostic and forensic applications4,5.
However, the adoption of protocols combining RT and qPCR necessitates acceptance of assumptions carried through from legacy RT-PCR; primarily that the process of RT is reproducibly quantitative, linear and maintains proportionality between genes when the cDNA for each is produced. It has become apparent that each of these assumptions requires further qualification before total adoption of any protocol is appropriate. In addition, the degree to which different protocols, RT enzymes and priming strategies influence quantitative assessments also requires clarification.
RT reaction components and protocols
Reverse transcriptases
A typical RT experiment requires the collection of tissue samples, RNA extraction and quality assessment, followed by cDNA synthesis. cDNA synthesis protocols are highly diverse and utilise different types of RNA-dependent DNA polymerase enzymes (reverse transcriptases). These occur naturally in some viruses, including retroviruses Avian Myeloblastoma Virus and Moloney Monkey Leukaemia Virus, which are the source for the commonly used RT enzymes, AMV or MMLV respectively. Both are characterised by low fidelity, with AMV having twice the mutation rate of MMLV. Contrary to popular belief, neither enzyme has an absolute requirement for a double stranded primer region for initiation of cDNA synthesis although priming serves to accelerate initiation. Since both process optimally at rather low temperatures, AMV at 42°C to 52°C and 37°C for MMLV, various modification have been introduced that have resulted in enzymes that function at higher temperatures (as high as 70°C). This helps to dissociate regions of secondary structure on RNA targets and allows reverse transcription of these stretches as well as those of high GC content. Some enzymes are not derived from MMLV or AMV-RT and are supposed to have a higher affinity for RNA than either. Omniscript RT is designed for reverse transcription of 50 ng-2 µg of RNA and is recommended for use with difficult-to-transcribe templates. The Sensiscript RT is designed for reverse transcription of less than 50 ng of RNA; it is not recommended for use with viral RNA when carrier RNA is present.
Commercial suppliers often provide blends of multiple enzymes to maximise the benefit from the best features of each. When reverse transcription is to be performed at high temperature, an initial lower temperature incubation is used to promote primer annealing. In addition, buffer compositions are also formulated to disrupt secondary structure by addition of e.g. 20% glycerol, 1M betaine or 2-10% DMSO or up to 4M sodium pyrophosphate. The different enzyme, buffer and RT temperature combination have dramatic effects on cDNA synthesis, resulting in up to 100-fold differences in cDNA yield of different targets6.
Priming strategies
There are several choices for cDNA priming strategies that involve reverse transcription from target-specific primers, oligo-dT and anchored variants thereof, various random priming procedures as well as combinations of the above.
- A specific primer targeting the RNA sequence can be used to direct RT towards the specific region of interest. In many cases this will be one of the PCR primers and conventional “one-step” protocols include a RT step with the reaction containing both primers. Priming with a target specific primer is particularly vulnerable to template secondary structure and so the assay design must include steps to ensure that the primer targets a region that is predicted to be open under the RT conditions determined for the primer7.
- Alternatively it may be desirable to produce cDNA that is representative of the entire RNA population. This is achieved by priming all transcripts using either a primer consisting of a string of thymidine residues (oligo dT) or collection of primers of random sequence (random primers). cDNA priming using olig-dT is critically dependent on RNA integrity, whereas random priming will reverse transcribe mostly rRNA and lose sensitivity. In theory oligo dT exclusively targets the 3′ polyA tract of mRNA although it is clear that it also hybridises to a rich regions of rRNA molecules. In order to increase specificity many oligo dT primers have two additional bases at the 3′ end (“anchored” primers). After hybridisation, RT proceeds from 3′ to 5′ although it is safest to assume that the RT will only capture the 3′ most 1200 bases. Alternatively a series of random sequences are hybridised along the length of all RNA molecules to prime cDNA fragments representative of the original RNA.
It has been widely described that the choice of priming strategies results in large differences in cDNA yield for given targets8. Many RT protocols call for an RNase H digestion step after cDNA synthesis. This is to remove the RNA strand since RNA:DNA hybrids are kinetically stronger than DNA:DNA and allow more efficient PCR primer binding during early PCR cycles. However, for qPCR it is unlikely that RNase H activity contributes significantly since the high initial denaturation steps are sufficient to melt the hybrids. Similarly avoidance of RNase H activity in RT enzymes is less of an issue when targeting short amplicons.
Reproducibility of the RT reaction
Ideally, replication of the RT process should yield identical data. However, it is clear that for identical samples processed under identical conditions, the cDNA yield for given target genes can be hugely variable between replicate cDNA synthesis reactions (see Figure 1). This would be of minimal concern if all targets were affected proportionally, but it has been observed that the relative proportion of targets is not always maintained. Specifically, those genes present at lower copy number (e.g. those typically of great interest) appear to be more vulnerable to cDNA yield concentration fluctuations, while those at higher copy number (e.g. often adopted as reference genes) are less variable (see Figure 1). Similar phenomena have also been observed when using oligo dT priming. For this reason it is important to validate the reproducibility of RT protocols, ensuring that protocols are designed such that all samples can be processed together or that an experimental calibrator is included so that batch-to-batch variations in the relative yield of each target can be corrected9.
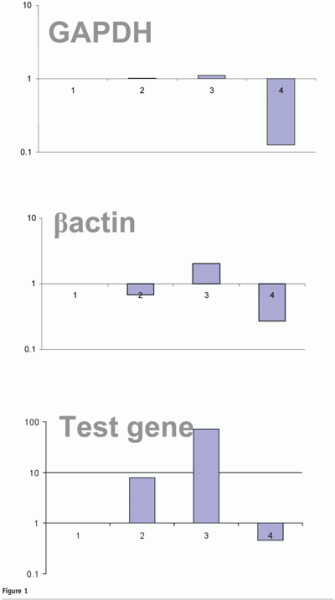

Linearity of RT reaction
In order to precede qPCR with RT there is an inherent assumption that the RT step is also quantitative. This can be tested by diluting RNA and after reverse transcription, using qPCR to measure the relative yield of a series of targets. However, when using protocols including either oligo dT or random primers the relative yield of cDNA is not directly proportional to the starting RNA concentration (see Figure 2). This phenomenon can be overcome by the addition of PEG8 and it is worthy of note that when a dilution series of RNA is primed with a gene specific primer the cDNA yield is directly proportional to the starting RNA concentration. It is important to validate that the RT protocol selected ensures a cDNA yield that is proportional to the input RNA or that all comparisons are of data referring to reactions containing equal concentrations of all RNA samples.
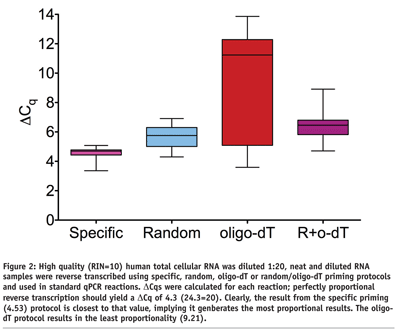

Reaction Inhibition
While reaction protocols result in highly variable data, reaction components may also contribute to inaccurate results. It has been clearly demonstrated that reverse transcriptase remains bound to cDNA templates and that this results in inhibition of the qPCR11. In addition to essential components of the RT-qPCR, reagents remaining from the sample storage and preparation procedure e.g. phenol, EDTA can also inhibit the reaction as well as those which derive from the sample which are not purified during RNA extraction e.g. urea or haem12. It is critical to examine the quality of samples to ensure that they do not contain substances that will inhibit the RT reaction13.
Conclusion
Consistency and reliability of RT-qPCR assays depend on the proper execution of a number of steps, in particular those relating to careful consideration of the quality of the RNA template, the choices of cDNA priming strategy and use of optimal reverse transcriptase. Not surprisingly, the pervasive penetration of this technology has led to the development of numerous, distinct and frequently divergent experimental protocols that often generate discordant results. Fortunately, the resulting uncertainty has increased the awareness of a need for common guidelines, in particular those relating to quality assessment of every component of the RT-qPCR assay and appropriate data analysis. This clear requirement for improved consistency of gene expression measurements is particularly relevant in relation to human clinical diagnostic assays and effective guideline regulating its use are essential for the future of molecular diagnostics within biomedical sciences. Detailed discussions of the many considerations essential for obtaining biologically relevant data can be found online (www.gene-quantification.info).
References
- Higuchi R, Dollinger G, Walsh PS, Griffith R. Simultaneous amplification and detection of specific DNA sequences. Biotechnology (NY) 1992;10:413-7.
- Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (NY) 1993;11:1026-30.
- Wittwer CT, Herrmann MG, Moss AA, Rasmussen RP. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques 1997;22:130-8.
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 2000;25:169-93.
- Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonak J, Lind K, et al. The real-time polymerase chain reaction. Mol Aspects Med 2006;27:95-125.
- Stahlberg A, Kubista M, Pfaffl M. Comparison of reverse transcriptases in gene expression analysis. Clin Chem 2004;50:1678-80.
- http://mfold.bioinfo.rpi.edu/
- Stahlberg A, Hakansson J, Xian X, Semb H, Kubista M. Properties of the reverse transcription reaction in mRNA quantification. Clin Chem 2004;50:509-15
- Lacey, HA, Nolan, T, Greenwood, SL, Glazier JD and Sibley CP. Gestational profile of Na+/H+ exchanger and Cl-/HCO3- anion exchanger mRNA expression in placenta using QPCR. Placenta 2005; 26, 93-98.
- Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nature Protocols 2006;1:1559-82.
- RT enzyme inhibition paper
- Huggett JF, Novak T, Garson JA, Green C, Morris-Jones SD, Miller RF, Zumla A. Differential susceptibility of PCR reactions to inhibitors: an important and unrecognised phenomenon. BMC Res Notes 2008;1:70.
- Stephen Bustin, Vladimir Benes, Jeremy Garson, Jan Hellemans, Jim Huggett, Mikael Kubista, Reinhold Mueller, Tania Nolan, Michael Pfaffl, Gregory Shipley, Jo Vandesompele, Carl Wittwer. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin Chem 2009 55:4




