Computational prediction of microRNA and targets
Posted: 29 September 2008 | | No comments yet
MicroRNAs (miRNAs) are small (~21nucleotides), evolutionarily conserved, noncoding RNA molecules that regulate gene expression1. In mammalian genomes, conservative predictions suggest that between 500-1500 miRNAs exist. There miRNAs appear to be capable of regulating the expression of multiple genes, with many genes appearing to be regulated by multiple, different, miRNAs2. Less conservative estimates suggest their may be tens of thousands of miRNAs3 in mammalian genomes, that between 20-30% of all human genes may be subject to regulation by miRNAs, and that each miRNA may contribute to the regulation of 200 or more mRNA targets4. Therefore it is easy to see why miRNA and their potential targets have received a lot of interest in recent times, as they offer a previously unknown mechanism of fundamental molecular biology that can subtly attenuate mRNA / protein expression.
For the pharmaceutical industry, the fact that miRNA have been associated with cancer5,6,7,8, neuroscience9,10,11,12, metabolism13,14 and the immune system15,16,17 has not gone unnoticed and has encouraged the appearance of new biotechnology companies which are actively pursuing miRNA therapeutic approaches for a number of different disease’s. Currently there are no direct miRNA therapeutics on the market, however similar approaches (i.e. antisense therapy) have been successfully launched and marketed, for example Fomivirsen made by Isis Pharmaceuticals, Inc. was approved by the FDA for the treatment of cytomegalovirus retinitis. It is therefore recognised that miRNA therapeutics offer great potential for drug discovery in the future.
Computational biology has been very much at the forefront of identifying and understanding the important roles that miRNA play as key gene regulators. Some of the fundamental computational approaches that have been employed to understand the molecular biology of miRNA will be discussed in this review, and some of the challenges facing researchers will be highlighted.
In silco target prediction
In 2003, it was shown that the Drosophila melanogaster miRNA Bantam targets and negatively regulates the pro-apoptotic gene hid18,19. This landmark event transformed the field of miRNA target prediction from a slow, complex, experimental approach to a sequence-based computational problem, with several groups publishing genome-wide methodology rapidly20,21,22,23. These algorithms generally calculate a score based on a measure of sequence homology between the query miRNA and the target sequence, they may also calculate the minimum free energy of the miRNA/target duplex and filter the sites based on the degree of conservation of the putative target site in orthologous genes.
Interestingly, these methods were based on very limited information regarding the biology and function of miRNAs. Firstly, only a very small set of experimentally confirmed miRNA/target sites were known, although this list is steadily growing. Secondly, an initial observation made by Lai et al suggested some regulatory motifs in the 3’ untranslated region (UTR) were perfectly complementary to the 5’ end of some fly miRNAs24. Thirdly, in vitro studies suggested that the multiplicity of miRNA sites in the 3’ UTR of genes seemed to increase the efficacy of target repression1. Based on this limited set of rules it is unsurprising that in silico target prediction is a less than perfect solution to a reasonably difficult computational problem. However, many of the algorithms predict functional miRNA/target interactions, but comparison at a genome scale reveals each algorithm predicts very different numbers of putative target sites with very little overlap25,26,27.
Generally miRNA target sites can be classified into three categories: i) 5’ dominant canonical, ii) 5’-dominant seed only and iii) 3’ compensatory28. The seed region is defined as a consecutive stretch of 6-8 nucleotides starting from the second nucleotide at the 5’ end of a miRNA (Figure 1). This region has been shown to be important with regard to guiding the silencing complex to its 3’UTR target1.
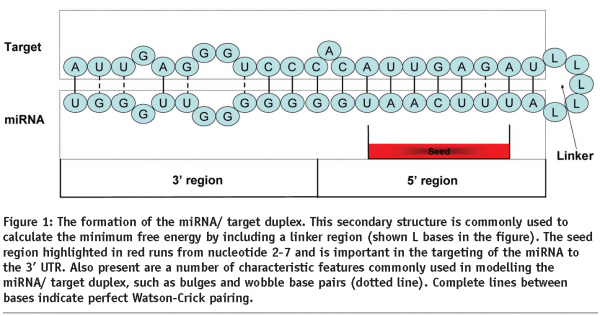

Canonical sites have perfect base pairing to at least the seed portion of the 5’ end of the miRNA and extensive base pairing to the 3’ end of the miRNA. The Seed only sites have perfect base pairing in the seed region only, but may have limited 3’ base pairing. Compensatory sites have less homology in the 5’ seed region which may include wobble pairs or mismatches, but have a greater degree of 3’ base pairing28.
miRanda
Given a microRNA sequence and 3’ UTR sequences, miRanda20,29 uses a dynamic programming alignment algorithm that locates all reasonably complementary sites for that miRNA in all given 3’ UTRs. Each potential site is evaluated for miRNA binding by calculating miRNA/target thermodynamics using the Vienna package30. The algorithm uses a scoring matrix that weights complementary bases at the 5’ end of the miRNA more than those at the 3’ end. This allows binding sites that have a perfect or near-perfect match to the seed region of miRNAs to have a better score20. A newer version of miRanda31 incorporates a statistical model equivalent to that used by RNAHybrid32 which allows miRanda to calculate a p value for each putative miRNA/target interaction, and to consider the multiplicity of miRNA sites in a single 3’ UTR and conservation across orthologous UTRs.
RNAhybrid
RNAhybrid32,33 was the first miRNA target prediction algorithm to approach the miRNA/target interaction from a thermodynamic/ RNA folding view point. The algorithm itself calculates the minimum free energy of potential miRNA / target binding sites, rather than using an artificial linker sequence (Figure 1) to force the miRNA / target into a suitable hairpin duplex as input into Mfold34 or the Vienna package30. This superior energy calculation coupled to a robust statistical frame work allowed RNAhybrid to address the fact that many of the prediction algorithms have a significant false positive rate, but as a user one could not distinguish between true positives and false positives. Therefore by modelling minimum free energies for a background distribution of randomly shuffled sequence, which approximates an extreme value distribution, it became possible to calculate a p value for any particular miRNA/target interaction. These p values supply the user with a degree of confidence as to how functional this interaction might be. Rehmsmeier et al then took the statistic a stage further and considered the effect of multiple target sites of the same miRNA within the same 3’ UTR. They reason that the occurrence of a single miRNA/target site would be classes as a rare event, and therefore multiple sites within the same 3’ UTR must be considered as very rare. This assumption and reasoning prompted the group to use the Poisson distribution as an approximation of the distribution of random multiple binding sites. This model was used to improve the base statistic, which is reflected as a more significant p value.
Both miRanda and RNAhybrid use a similar statistical frame work, however they approach the scoring of potential miRNA/target duplex’s from fundamentally different view points. The putative miRNA/target sites from both of the algorithms are available from the following website’s; (http://microrna.sanger.ac.uk/targets/ and http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/ ), but more importantly can be downloaded and run as local programs with your own miRNA sequences and 3’ UTRs , unlike TargetScan23,35 or PicTar21 two other highly used and cited algorithms.
Prediction of miRNA Genes
As with miRNA target prediction, miRNA gene prediction has largely been looked on as computational problem. As more miRNAs have been discovered certain features of the miRNA primary transcripts have become more apparent, which has then aided further rounds of algorithm development. For instance miRNA are transcribed from the genome as a single or polycistronic message, which then folds into characteristic stem-loop secondary structures. This characteristic stem-loop structure is a well established feature of miRNA, as the biogenesis of the mature miRNA requires the hairpin structure, which is then processed by drosha and dicer (for review see36).
Saini et al37 used a systematic approach to consider multiple sets of transcriptional features from all known human intergenic miRNAs. These features such as the transcription start sites, CpG island, the location of gene identification signature-paired-end ditags, transcription factor binding site location and the presence of poly(A) signals were used to show that for intergenic miRNA a significant proportion of primary transcripts are 3-4 kb in length. Not only does this indicate that primary miRNA transcripts are long, but they also possess features commonly associated with promoter regions of coding transcripts. For those miRNA that are located in the intron of host genes, it is widely considered that they are transcribed and coexpressed as part of their host genes pre-mRNA, and after splicing form the characteristic hairpin structures.
A basic general protocol for identifying miRNA genes could start by using BLAST38 and comparing all known miRNAs available from the miRBase sequence repository (http://microrna.sanger.ac.uk) against the genome of choice. Unfortunately, the statistics for BLAST are not really suitable for such short sequences, however BLAST is only used at this stage as a preliminary analysis filter to identify candidate regions similar to known miRNAs. The next step is to take sequence from approximately 100 bps up and down stream of the candidate regions identified by the sequence search. These sequences are then folded using a secondary structure prediction program such as Mfold34 or the Vienna package30. Candidates regions which form hairpin structures with a low minimum free energy and have homology with other known miRNAs should be taken on and validated in vitro. If the regions are confirmed as expressed and function as miRNA, the sequence should be registered at miRBase2 where the new miRNA will receive its official name prior to publication.
A major weakness in the above approach is caused by the initial sequence search being based on identifying regions in the genome that show homology to other known miRNA. This limitation of searching for miRNA like those already known could make the approach insensitive to finding truly novel miRNA. In order to approach the issue of lack of novelty, the use of phylogenetic shadowing has been used successfully, and has identified a number of experimentally confirmed novel miRNA2,5,39.
Other methods such as RNA223 use a pattern based approach for finding miRNA binding sites and identifying corresponding miRNA sequences. The RNA22 algorithm systematically assesses every potential binding site pattern in the given genome. Statistically significant candidate binding sites are then reverse complemented, and their location found in the genome. By using this process not only are the target sites found, but also the miRNA genes that interact with them. The author’s results indicate that in human and mouse >25,000 precursor miRNA can be identified3. This number seems incredibly high, however data from massively parallel sequencing carried out in different cell types, is starting to suggest that we initially underestimated the number of miRNAs present in mammalian genomes. For example in pluripotent human embryonic stem cells 83 novel miRNA were identified using next generation sequencing40, in human and chimpanzee brain an additional 447 novel miRNA have been identified9. Therefore the number of miRNA genes is definitely expected to continue increasing in the near future as the use of next generation sequencing is used to profile different tissues / cells and disease states.
Systems Biology of miRNA Regulation
One of the current challenges facing the miRNA field is dealing with, and understanding, the product of genome-scale systematic screening or profiling of miRNA. Many of the microarray technology companies now offer miRNA microarray chips, also with the arrival of next generation sequencing technology, the ability to rapidly and systematically profile the expression of miRNA in experiments has arrived. However, it is still very difficult to i) handle the vast quantities of data produced and ii) use the generated profiles to establish which mRNAs / genes are regulated by the miRNAs present.
One approach is to use the computational methodologies described above to assess if the miRNAs identified in the profile have any putative targets in the genome / tissue / cell line of interest. However given the nature of the prediction algorithms one has to be careful when doing this, prediction algorithms will give you an answer, however it might not be the correct one.
Therefore a number of publications have approached this problem from a miRNA seed enrichment perspective using transcriptomic data. If one considers only direct interactions between miRNA and target mRNA it is possible to make sense of some large scale experimental data. Two examples of enrichment analysis have been published, one in a transgenic zebrafish (Danio rerio) which has had miR-430 knocked out41 and the other, a BIC/miR-155 knockout mouse16.
In both of these publications a transcriptomic experiment was carried out on a standard gene chip. Differentially expressed genes were obtained via comparison between knockout and wild type individuals, resulting in a list of more than 328 mRNAs being up regulated at significant levels (≥1.5-fold, p≤0.05) in the miR-430 knockout41, and ~ 100 mRNAs up regulated in the miR-155 knockout mouse16. The 3’ UTRs of these genes were then mined for miRNA seed regions and compared back to the count of those seed regions present in the genome as a whole. For confidence, a simple Fishers exact test42 can be used to assess significance, or fold-enrichment can be calculated to identify enriched miRNA seed16. These publications show enrichment for seed regions from miRNA which were initially knocked out (miR-430 and miR-155) in the original organism. This result would be expected, as one assumes that miRNA are repressors, and therefore the up regulated genes identified in the knockout verses the wild type, are those that are influenced by the presence / absence of the miRNA.
Ideally, these two approaches highlighted above should be complementary, in that genes that are down regulated in the presence of a miRNA identified in miRNA profile should show enrichment for the seed region of that miRNA. Also the in silico prediction methods should identify putative miRNA/target sites within the 3’UTRs of those down regulated genes.
The main caveat with these approaches is that they are limited to making sense of only the direct miRNA/target interaction, and offers no way of making sense of indirect up and down stream gene interactions. Also these approaches are only applicable if the miRNA is acting at the transcriptional level, as miRNA influencing protein translation, such as miR-146a43, will have little to no direct effect in a transcriptomic experiment. Analysis of the anomalies identified using such approaches my yield important to our knowledge and may enhance further roles of algorithm development.
The issue of deconvoluting direct and indirect interactions is fundamental in understanding what the effect of a particular miRNA is. Also it is important when considering how that miRNAs influence relates to the cellular level and / or the pathway level. The deconvolution of miRNA interaction networks gained from large scale genomic screening is a difficult problem which needs to be solved before we can truly see the full influence of miRNA.
References
- Bartel, D. P.,2004.MicroRNAs: Genomics, Biogenesis, Mechanism, and Function.Cell.116,281-297.
- Griffiths-Jones, S., Grocock, R. J., Van Dongen, S., Bateman, A., and Enright, A. J.,2006.MiRBase: MicroRNA Sequences, Targets and Gene Nomenclature.Nucleic Acids Res.34,D140-D144.
- Miranda, K. C., Huynh, T., Tay, Y., Ang, Y. S., Tam, W. L., Thomson, A. M., Lim, B., and Rigoutsos, I.,2006.A Pattern-Based Method for the Identification of MicroRNA Binding Sites and Their Corresponding Heteroduplexes.Cell.126,1203-1217.
- Lim, L. P., Lau, N. C., Garrett-Engele, P., Grimson, A., Schelter, J. M., Castle, J., Bartel, D. P., Linsley, P. S., and Johnson, J. M.,2005.Microarray Analysis Shows That Some MicroRNAs Downregulate Large Numbers of Target MRNAs.Nature.433,769-773.
- Berezikov, E. and Plasterk, R. H.,2005.Camels and Zebrafish, Viruses and Cancer: a MicroRNA Update.Hum.Mol.Genet.14 Spec No. 2,R183-R190.
- Calin, G. A. and Croce, C. M.,2006.MicroRNAs and Chromosomal Abnormalities in Cancer Cells.Oncogene.25,6202-6210.
- Dalmay, T. and Edwards, D. R.,2006.MicroRNAs and the Hallmarks of Cancer.Oncogene.25,6170-6175.
- Fabbri, M., Croce, C. M., and Calin, G. A.,2008.MicroRNAs.Cancer J.14,1-6.
- Berezikov, E., Thuemmler, F., van Laake, L. W., Kondova, I., Bontrop, R., Cuppen, E., and Plasterk, R. H.,2006.Diversity of MicroRNAs in Human and Chimpanzee Brain.Nat.Genet.38,1375-1377.
- Cogswell, J. P., Ward, J., Taylor, I. A., Waters, M., Shi, Y., Cannon, B., Kelnar, K., Kemppainen, J., Brown, D., Chen, C., Prinjha, R. K., Richardson, J. C., Saunders, A. M., Roses, A. D., and Richards, C. A.,2008.Identification of MiRNA Changes in Alzheimer’s Disease Brain and CSF Yields Putative Biomarkers and Insights into Disease Pathways.J.Alzheimers.Dis.14,27-41.
- Giraldez, A. J., Cinalli, R. M., Glasner, M. E., Enright, A. J., Thomson, J. M., Baskerville, S., Hammond, S. M., Bartel, D. P., and Schier, A. F.,2005.MicroRNAs Regulate Brain Morphogenesis in Zebrafish.Science.308, 833-838.
- Miska, E. A., Alvarez-Saavedra, E., Townsend, M., Yoshii, A., Sestan, N., Rakic, P., Constantine-Paton, M., and Horvitz, H. R.,2004.Microarray Analysis of MicroRNA Expression in the Developing Mammalian Brain.Genome Biol.5,R68-
- Krutzfeldt, J. and Stoffel, M.,2006.MicroRNAs: a New Class of Regulatory Genes Affecting Metabolism.Cell Metab.4,9-12.
- Wilfred, B. R., Wang, W. X., and Nelson, P. T.,2007.Energizing MiRNA Research: a Review of the Role of MiRNAs in Lipid Metabolism, With a Prediction That MiR-103/107 Regulates Human Metabolic Pathways.Mol.Genet.Metab.91,209-217.
- Baltimore, D., Boldin, M. P., O’Connell, R. M., Rao, D. S., and Taganov, K. D.,2008.MicroRNAs: New Regulators of Immune Cell Development and Function.Nat.Immunol.9,839-845.
- Rodriguez, A., Vigorito, E., Clare, S., Warren, M. V., Couttet, P., Soond, D. R., Van Dongen, S., Grocock, R. J., Das, P. P., Miska, E. A., Vetrie, D., Okkenhaug, K., Enright, A. J., Dougan, G., Turner, M., and Bradley, A.,2007.Requirement of Bic/MicroRNA-155 for Normal Immune Function.Science.316,608-611.
- Taganov, K. D., Boldin, M. P., Chang, K. J., and Baltimore, D.,2006.NF-KappaB-Dependent Induction of MicroRNA MiR-146, an Inhibitor Targeted to Signaling Proteins of Innate Immune Responses.Proc.Natl.Acad.Sci. U.S.A.103,12481-12486.
- Brennecke, J., Hipfner, D. R., Stark, A., Russell, R. B., and Cohen, S. M.,2003.Bantam Encodes a Developmentally Regulated MicroRNA That Controls Cell Proliferation and Regulates the Proapoptotic Gene Hid in Drosophila.Cell.113,25-36.
- Stark, A., Brennecke, J., Russell, R. B., and Cohen, S. M.,2003.Identification of Drosophila MicroRNA Targets.PLoS Biol.1,E60-
- Enright, A. J., John, B., Gaul, U., Tuschl, T., Sander, C., and Marks, D. S.,2003.MicroRNA Targets in Drosophila.Genome Biol.5,R1-
- Rajewsky, N. and Socci, N. D.,2004.Computational Identification of MicroRNA Targets.Dev Biol.267,529-35.
- Kiriakidou, M., Nelson, P. T., Kouranov, A., Fitziev, P., Bouyioukos, C., Mourelatos, Z., and Hatzigeorgiou, A.,2004.A Combined Computational-Experimental Approach Predicts Human MicroRNA Targets.Genes Dev.18,1165-78.
- Lewis, B. P., Shih, I. H., Jones-Rhoades, M. W., Bartel, D. P., and Burge, C. B.,2003.Prediction of Mammalian MicroRNA Targets.Cell.115,787-98.
- Lai, E. C.,2002.Micro RNAs Are Complementary to 3′ UTR Sequence Motifs That Mediate Negative Post-Transcriptional Regulation.Nat Genet.30,363-4.
- Barnes, M. R., Deharo, S., Grocock, R. J., Brown, J. R., and Sanseau, P.,2007.The Micro RNA Target Paradigm: a Fundamental and Polymorphic Control Layer of Cellular Expression.Expert Opin.Biol.Ther.7,1387-1399.
- Maziere, P. and Enright, A. J.,2007.Prediction of MicroRNA Targets.Drug Discov.Today.12,452-458.
- Rajewsky, N.,2006.MicroRNA Target Predictions in Animals.Nat.Genet. 38 Suppl,S8-13.
- Brennecke, J., Stark, A., Russell, R. B., and Cohen, S. M.,2005.Principles of MicroRNA-Target Recognition.PLoS.Biol.3,e85-
- John, B., Enright, A. J., Aravin, A., Tuschl, T., Sander, C., and Marks, D. S.,2004.Human MicroRNA Targets.PLoS Biol.2,e363-
- Wuchty, S., Fontana, W., Hofacker, I. L., and Schuster, P.,1999.Complete Suboptimal Folding of RNA and the Stability of Secondary Structures.Biopolymers.49,145-165.
- Griffiths-Jones, S., Grocock, R. J., Van Dongen, S., Bateman, A., and Enright, A. J.,2006.MiRBase: MicroRNA Sequences, Targets and Gene Nomenclature.Nucleic Acids Res.34,D140-D144.
- Rehmsmeier, M., Steffen, P., Hochsmann, M., and Giegerich, R.,2004.Fast and Effective Prediction of MicroRNA/Target Duplexes.RNA.10,1507-1517.
- Kruger, J. and Rehmsmeier, M.,2006.RNAhybrid: MicroRNA Target Prediction Easy, Fast and Flexible.Nucleic Acids Res.34,W451-W454.
- Zuker, M.,2003.Mfold Web Server for Nucleic Acid Folding and Hybridization Prediction.Nucleic Acids Res.31,3406-3415.
- Lewis, B. P., Burge, C. B., and Bartel, D. P.,2005.Conserved Seed Pairing, Often Flanked by Adenosines, Indicates That Thousands of Human Genes Are MicroRNA Targets.Cell.120,15-20.
- He, L. and Hannon, G. J.,2004.MicroRNAs: Small RNAs With a Big Role in Gene Regulation.Nat.Rev.Genet.5,522-531.
- Saini, H. K., Griffiths-Jones, S., and Enright, A. J.,2007.Genomic Analysis of Human MicroRNA Transcripts.Proc.Natl.Acad.Sci.U.S.A.104, 17719-17724.
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J.,1990.Basic Local Alignment Search Tool.J.Mol.Biol.215,403-410.
- Berezikov, E., Guryev, V., van de, Belt J., Wienholds, E., Plasterk, R. H., and Cuppen, E.,2005.Phylogenetic Shadowing and Computational Identification of Human MicroRNA Genes.Cell.120,21-24.
- Morin, R. D., O’Connor, M. D., Griffith, M., Kuchenbauer, F., Delaney, A., Prabhu, A. L., Zhao, Y., McDonald, H., Zeng, T., Hirst, M., Eaves, C. J., and Marra, M. A.,2008.Application of Massively Parallel Sequencing to MicroRNA Profiling and Discovery in Human Embryonic Stem Cells.Genome Res.18,610-621.
- Giraldez, A. J., Mishima, Y., Rihel, J., Grocock, R. J., Van Dongen, S., Inoue, K., Enright, A. J., and Schier, A. F.,2006.Zebrafish MiR-430 Promotes Deadenylation and Clearance of Maternal MRNAs.Science.312,75-79.
- Fisher, R.,1922.On the Interpretation of Chi Squared From Contingency Tables, and the Calculation of p.Journal of the Royal Statistical Society.85,87-94.
- Perry, M. M., Moschos, S. A., Williams, A. E., Shepherd, N. J., Larner-Svensson, H. M., and Lindsay, M. A.,2008.Rapid Changes in MicroRNA-146a Expression Negatively Regulate the IL-1beta-Induced Inflammatory Response in Human Lung Alveolar Epithelial Cells.J.Immunol.180,5689-5698.