Tracking and tracing to the root cause: case studies in microbial contamination
Posted: 11 April 2022 | Tim Sandle (Bio Products Laboratory) | 2 comments
In this article, pharmaceutical microbiologist and contamination control expert Tim Sandle presents three microbial contamination investigation case studies, highlighting the key lessons for pharmaceutical microbiologists to take away and the underlying importance of identifying the root cause of microbial data deviations.

Introduction
One of the main activities of the pharmaceutical microbiologist is with determining the contamination control strategy and proactively identifying measures to lower identified risks and, where risks remain of concern, to introduce monitoring. However, there will invariably be microbial recovery, from product (intermediate and finished), from water, cleanrooms and other utilities. A common term of these events is ‘microbial data deviations’, although other terminology can apply.1 These microbial events require investigation and the importance of such an investigation is elevated where there is recurrence. While investigations are referred to in regulatory documents, there is a dearth of case studies to help guide microbiologists and Quality Assurance departments. This article presents three case studies. While the specific issues may or may not be of direct relevance, the areas examined and the thought processes will be of wider applicability.
Microbial investigations
Microbial data deviations can be categorised as:
- Out of limits results (OOL), which is often applied to environmental monitoring
- Out of specification results (OOS), which is typically used for pharmacopeia tests
- Out of trend (OOT), as defined in relation to a trend chart.
Microbial data deviations need to form part of the quality system and they require an investigation. The target time for microbiology investigations is to be completed within thirty days, against a procedure.
Meet Tim Sandle at The Future of Bio/Pharmaceutical Analysis Online Summit this May!
Join Tim’s Microbiology Masterclass to discover the techniques simplifying and expediting microbiology workflows and learn how to ensure your contamination control strategy performs at its best.


The optimal way to gather evidence is to visit the area, where the role of the pharmaceutical microbiologist is mostly in the process area.2 In addition, operators may need to be interviewed and the information should be captured and documented. A further important source of information can be the batch manufacturing or batch processing record. Visits and document reviews can be supported by additional sampling. With sampling, it is best practice to have a hypothesis developed in advance together with the numbers and types of samples detailed. More than one sampling session may be required to account for the variances with microbial distribution.3 Further evidence should be gathered from accurate microbial species identification.
The three case studies draw on these essential elements and demonstrate how things can sometimes go wrong.
Case study 1: Microbial contamination in non-sterile manufacturing
The first scenario relates to tablet manufacture. The company in question noted repeated finished product failures with a Bacillus species, human skin commensals and Gram-negative rods. Individual tablets had a bioburden estimated to be more than 2,500 colony forming units (CFU), containing the mixed flora.
There was no obvious pattern in relation to the contamination, which was found throughout the batch. Further examinations indicated that multiple batches were found to be contaminated. The important starting point was to undertake microbial identifications to species level, as understanding the microorganisms can provide clues as to the habitat and possible origin. The pattern of different microorganisms also helps with comparing microorganisms isolated in the finished product with organisms recovered from other sources, such as water and the cleanroom, and to track contamination at different points in the process.
The main part of the investigation required a construction of a process flow chart, to enable the manufacturing process to be examined.
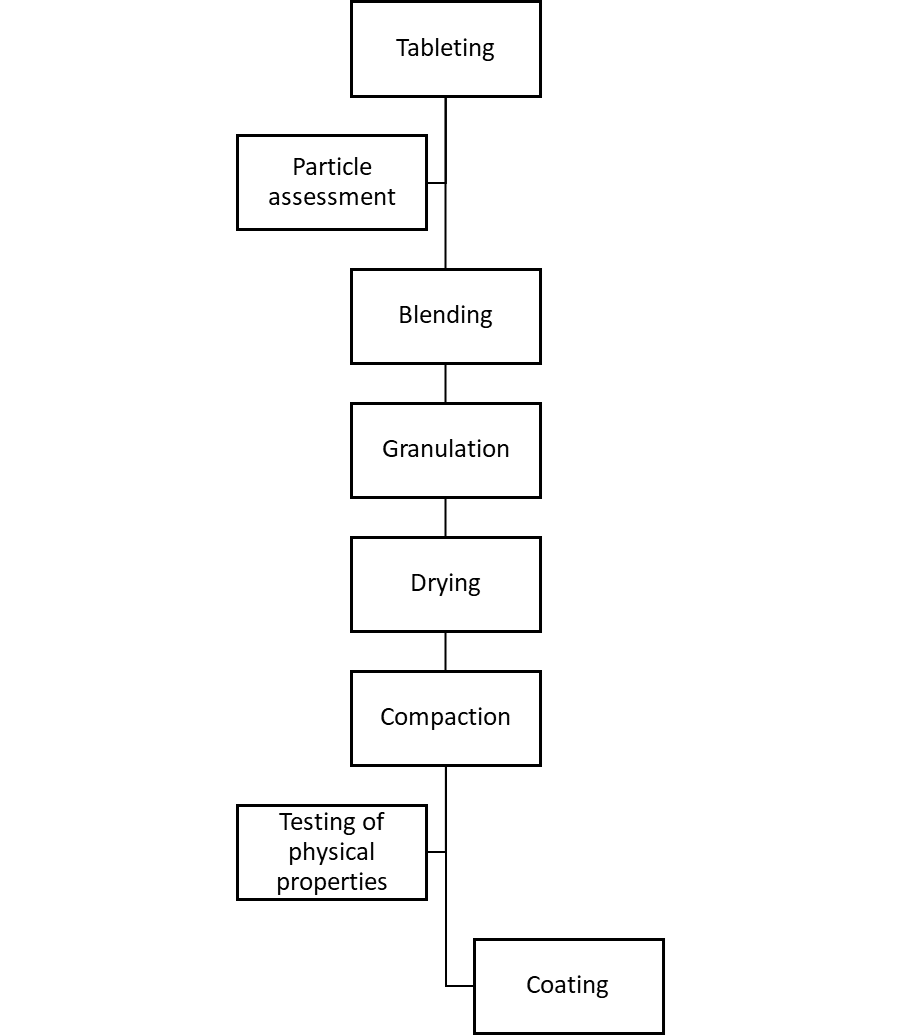

Figure 1: General steps for tablet manufacturing.
The main manufacturing steps, together with initial observations, were:
- Wet mixing of raw materials – this is performed with purified water. The successful mixing of the powder is more difficult than mixing liquid and is based on achieving homogeneity. The ease of this is dependent upon the inherent cohesiveness and resistance to movement between the individual particles.
- Drying – with drying, it is important to keep the residual moisture low enough to prevent product deterioration and ensure free flowing properties. The drying process was found to leave the product as a cake with a high level of residual humidity within it.
- Bulking up – the bulking up process allowed multiple cakes to be transported to the tanks and added via a vacuum lance.
- Compression – compression was believed by the company to be antimicrobial. This was based on development work that provided evidence of a multi-log reduction in the microbial population.
- Packing – the packing process was into blisters, taking place within a controlled environment.
In addition, environmental monitoring data was examined. Here it was found that the routine monitoring of the manufacturing room had not recovered anything atypical. As well as the cleanroom, the purified water system had not seen any recent recoveries of microorganisms.


For the next stage of the investigation, the equipment in the manufacturing process was opened up and sampled microbiologically. In addition, testing was undertaken of the raw material, a substance of natural origin that was found to contain a low level of Gram-negative bacteria. It was of interest that these organisms were of the type seen in the failed batches, but the numbers recovered were relatively low. No organisms were recovered at the wet mixing stage.
The investigation proceeded to the drying stage, looking at the room equipment. One area examined, the extract pipework from the dryer, was found to have pools of condensate inside it. Due to a process improvement step, the extract was no longer being used due to the discontinuation of a product line. A previous version of the product used alcohol rather than water as the mixing agent (the extract was a safety precaution to remove the alcohol fumes). Additional testing revealed that the extract condensate was heavily contaminated with Gram-negative bacteria.
For the investigation of the bulking up stage, the operation of the lance was found to be satisfactory. However, increased environmental monitoring was undertaken and the active (volumetric) air samples taken within the lancing area cleanroom recovered a combination of Bacillus species and skin commensals. Other areas examined included:
- The tank – opened up via the main ‘lid’, further inspection was difficult because there was no manway
- The connection from tank wall to tank lid, which formed a lip about 10cm wide. The lip was covered in a old grey This residual powder was heavily contaminated with the microorganisms isolated from the finished tests.
- The lid of the tank, which had evidence of condensate on it.
- The room HEPA inputs – based on the walls (rather than being ceiling mounted, which would be more conventional), with cold filtered air continuously bathed the top of the tank.
With the study of the compression and packing lines, these recovered no microbial bioburden.
A further important element of any microbial investigation is with the trend. This is examined by constructing a trend chart and overlaying historical events. The trend analysis showed that the finished product failures, due to microbial contamination, began appearing after the alcohol containing product was discontinued.
The overall findings demonstrated a set of different failure modes:
- The wet cake enabled the survival of Gram-negative bacteria
- The lance activity pulled human skin and environmental isolates into the tank
- The old air trapped in the tank allowed condensate to form and
- the poor design of the tank retained the condensate, allowing it to hit the joining tank lip.
- The lip allowed contaminated powder to accumulate. The powder provided a nutritive source which facilitated bacteria to grow.
The reason that the issue appeared, and had not been noted previously, was because the discontinued alcohol containing product would have sanitised the filling equipment. In addition, the compression unit would have helped to have eliminated any low-level microorganisms that would have survived the alcohol step. The removal of the alcohol highlighted the poor controls through the process and led to conditions that enabled the proliferation of microorganisms.
Identifying these areas enabled appropriate corrective and preventative actions to be set. In particular, plant modifications and routine cleaning was introduced.
Case study 2: Environmental monitoring – the unexplained organism
The second case study, relates to a manufacturing operation within a Grade C cleanroom. From an environmental monitoring session, a working height surface contact plate of a conveyor belt recovered a high level of bacteria, with the plate being ‘too numerous to count’ (against an action level of 25 CFU/cm2). As was customary, the isolates were Gram-stained and then a phenotypic identification was performed using a semi-automated commercial system. The predominant isolate was Yersinia pestis. Other isolates were a set of more typical skin biota of the Staphylococcus species.


The laboratory analyst did not question the Y. pestis identification and the importance of the organism was realised by the supervisor. This is a facultative anaerobic organism that can infect humans via the Oriental rat flea, resulting in plague.4
The supervisor notified site management that ‘plague’ had been detected at the facility and unsurprisingly a state of panic ensued. Among the activities seriously being considered were site evacuation, notification of local authorities and placing everyone in the microbiology department under enforced isolation.
A more experienced microbiologist was called in and began assembling the facts. The microbiologist determined:
- The recovery of ‘bubonic plague’ from a conveyer belt did not make sense
- The environmental monitoring was performed on night shift
- Post incubation, the plate count was also performed on a night shift
- The microbial identification, following subculture, was also performed on night shift
- The supervisory review and subsequent alert of the discovery of plague was communicated on the night
The reason why the identification was most likely to be inaccurate was quickly established by:
- The apparent pestis was growing on the environmental monitoring agar, which was tryptone soya agar (TSA). In practice, pestis is isolated on stained smears of peripheral blood, lymph node specimens or from sputum. Y. pestis will grow as small, non- lactose fermenting colonies on MacConkey agar or Enterobacteriaceae selective agar, like EMB agar. These are supported by rapid microbiology assays.5
- The particular colony did not look morphologically like a Gram-negative. Although there are limitations with visual identification, the colonial morphology of Gram-negative and Gram-positive bacteria can often be distinctive.
- A repeat microscopic investigation, requested by the experienced microbiologist, of the older original colony showed the presence of endospores, which pestis does not form.
The bacterium was not Y. pestis. The microbiologist established that the errors that led to the misidentification were:
- The original Gram stain was incorrect – a young colony was stained and the outcome was Gram-variable; however, the analyst selected Gram-negative as the outcome
- The incorrect Gram stain led to the selection of the incorrect identification biochemical test kit
- The bacteria still reacted or grew within a particular patter of biochemical wells, providing a pattern that that was closely matched to a database and the most probable result selected by the software. This was indicated to be pestis.
Rather than questioning the oddness of the result, an emotive response from the supervisor led to rapid escalation of the identification result and considerable business disruption.
From a human factors perspective, the night shift is not a popular timeslot for more experienced microbiologists to work, leading to less experienced personnel often being allocated. Furthermore, personnel are often tired and sometimes fatigue adds to the judgement process.
Case study 3: Sterile product out of specification
The third and final case study relates to a sterile active pharmaceutical ingredient (API) facility who manufacture an injectable dry powder. The product is sterile filtered into a pre-sterilised manufacturing line and then turned into a dry powder and offloaded via Grade A isolators.
With one batch, there was a subsequent sterility test failure. This led to the recovery of two different Bacillus species and an OOS investigation. During the first phase of the investigation, there was a second sterility test failure with the same type of product. This batch also recovered the same two Bacillus species, expanding the scope of the investigation.
The investigation was split into two parts:6 the first looked at laboratory error and the second at the potential for a manufacturing failure (based on the typical first two phases of the OOS process). Across both parts, environmental monitoring was deployed to extensively sample manufacturing cleanroom air and surfaces; laboratory air and surfaces; isolator air and surfaces; and samples were taken from utilities like compressed air, nitrogen, water-for-injection (WFI), purified water, glycol, plant steam, heating ventilation and air conditioning (HVAC) ducting. A few environmental samples were also taken from support areas, such as the warehouses and controlled-non-classified (CNC) areas.

The environmental monitoring looked at the traditional techniques such as contacts and air samples. Some techniques had to be modified in order to allow for the sampling of air systems, glycol lines and high-pressure steam. The focus of the monitoring was less concerned with the numbers of organisms recovered and more with the types of microorganisms recovered, in an attempt to see if the two Bacillus species could be identified.
Many of the environmental monitoring samples recovered Bacillus in numerous places. The internal phenotypic identification system isolated the two specific species from the laboratory samples and one of the species from a production compressed air sample. The organisation concluded the results were a false positive based on finding the isolates in the laboratory, supported by the fact that two species had not been isolated together from the manufacturing plant. This was reported to senior management.
However, the isolates had also been sent away to an external laboratory for genotypic matching. The genotypic test results revealed that although the species of Bacillus were similar, the specific strains of the Bacillus organisms had not been found in the laboratory. In contrast, the species of Bacillus from the compressed air was genetically related and thus the same strain as the contaminant isolated from the two sterility test failures.
The dilemma was that the original phenotypic results had been reported with the point of origin directed to the laboratory, whereas the definitive data was now pointing towards a production related issue (the compressed air). The recommendation was made that the batches needed to be rejected, although the first reporting of the erroneous root cause caused considerable disagreement in senior management circles.
Lessons to be learned?
In terms of what can be drawn from these three case studies, environmental sampling and monitoring is not only about the numbers of organisms recovered; the type of species isolated can help with establishing origins and trending. The species need to be sense checked: isolates thus far only found in marine trenches or on the rim of volcanoes are unlikely to be recovered from cleanrooms. There is also a need, when batch disposition is resting on the identification, to draw on genotyping beyond the more conventional biochemical or proteomic techniques. An additional area where environmental monitoring can be useful is with moving sampling beyond the routine (to demonstrate a cleanroom remains in compliance); it can also serve as a powerful investigative tool to aid a deeper dive into investigations. This may include additional samples or sampling from areas that are not ordinarily assessed.
Summary
The case studies outlined are based on real-life events, collected across the pharmaceutical industry by the Pharmaceutical Microbiology Interest Group.7 In reading such events there may be a sense of ‘this would not happen to me’; however, it is a recurrent criticism from regulatory inspectors that many investigations are not sufficiently thorough or fail to get to the actual root cause. These case studies may provide advice that can be applied to an organisation’s microbial investigation procedure.
About the author
Dr Tim Sandle has over 25 years’ experience of microbiological research and biopharmaceutical processing. Tim is a member of several editorial boards and has authored 30 books on microbiology, healthcare and pharmaceutical sciences. Tim works for Bio Products Laboratory Limited (BPL) in the UK and is a visiting tutor at both the University of Manchester and UCL.
References
- McCullough K, Moldenhauer J. (2015) Introduction in Microbial Risk and Investigations. In McCullough, K. and Moldenhauer, J., (Eds) Microbial Risk and Investigations, Parenteral Drug Association (PDA) and Davis Healthcare International (DHI). Bethesda, MD, pp1-10
- Sutton S. (2011) Successful Microbial Investigations, American Pharmaceutical Review, 74 (2): 34-42
- Sandle T. (2014) Data Review and Analysis for Pharmaceutical Microbiology, Microbiology Solutions, UK., pp1-15
- Deng W, Burland V, Plunkett G, et al. (2002) Genome sequence of Yersinia pestis. Journal of Bacteriology. 184 (16): 4601–11
- Chanteau S, Rahalison L, Ralafiarisoa L, et al. (2003) Development and testing of a rapid diagnostic test for bubonic and pneumonic plague. Lancet 361: 211-216
- Sandle T. (2012): Sterility Test Failure Investigations, Journal of GxP Compliance, 16 (1): 1- 10
- Keen D. (2019) Contamination Case Studies, presented at Pharmig Irish Conference, 25th May 2019
If you enjoyed this article, we’re sure you’ll enjoy our sessions at The Future of Bio/Pharmaceutical Analysis Online Summit!
Join and interact with thought leaders as they present emerging research, discuss critical areas of technological advancement, as well as advise on analytical quality by design and regulatory and pharmacopoeial guidance for techniques spanning spectroscopy, chromatography, electrophoresis and more across our two-day programme.







What if no root cause/probable cause found in microbial investigation in non-sterile product?
This is very informative, lots of useful information presented here.