Affinity-based screening
Posted: 20 July 2006 | | No comments yet
Drug (lead) discovery relies on massive screening of chemical libraries against various extra- and intracellular molecular targets to find compounds with the desired mode of action. Sequencing of the human genome1 has generated a large number (>40 per cent) of new molecular targets with unknown function (‘orphan targets’), as well as a large number of molecular targets with known function albeit non-tractable by standard high-throughput screening (HTS) due to the particular requirements of plate-based assays in robotic screening systems (‘non-tractable targets’). Examples of the latter are targets with very fast kinetics, targets with multiple modes of function for the same polypeptide chain or targets where the substrate of a particular enzyme is not known.
Drug (lead) discovery relies on massive screening of chemical libraries against various extra- and intracellular molecular targets to find compounds with the desired mode of action. Sequencing of the human genome1 has generated a large number (>40 per cent) of new molecular targets with unknown function (‘orphan targets’), as well as a large number of molecular targets with known function albeit non-tractable by standard high-throughput screening (HTS) due to the particular requirements of plate-based assays in robotic screening systems (‘non-tractable targets’). Examples of the latter are targets with very fast kinetics, targets with multiple modes of function for the same polypeptide chain or targets where the substrate of a particular enzyme is not known.
Drug (lead) discovery relies on massive screening of chemical libraries against various extra- and intracellular molecular targets to find compounds with the desired mode of action. Sequencing of the human genome1 has generated a large number (>40 per cent) of new molecular targets with unknown function (‘orphan targets’), as well as a large number of molecular targets with known function albeit non-tractable by standard high-throughput screening (HTS) due to the particular requirements of plate-based assays in robotic screening systems (‘non-tractable targets’). Examples of the latter are targets with very fast kinetics, targets with multiple modes of function for the same polypeptide chain or targets where the substrate of a particular enzyme is not known.
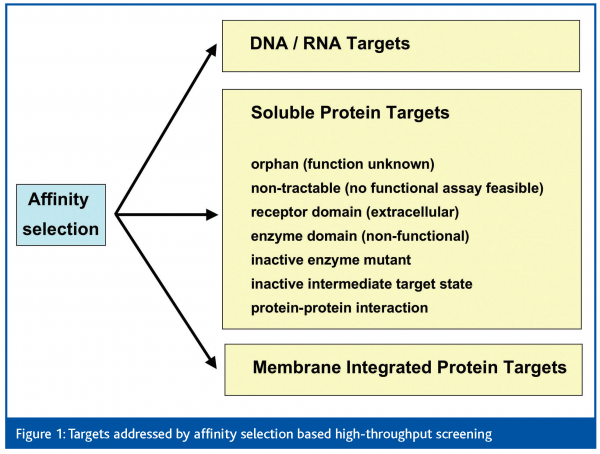
All together, this raises the question: to what extent and by which technology can orphan and non-tractable targets be approached in lead discovery, by means of HTS? As both types of targets are not amenable to functional assays in high throughput mode, affinity-based screening is the most promising way forward to search for molecules with a potential to bind and modulate the function of a particular target. It can be assumed that any ligand (binder) that interferes with the biological function of a target needs to bind to that target in a specific manner. Despite specific binding, also low-affinity and often non-specific interaction of a ligand with the target is possible. Whether binding occurs at an active or allosteric site of an enzyme, or at an agonistic or antagonistic receptor binding site, is an open issue and must often be characterised by appropriate secondary assays. Moreover, upon binding of a ligand (or prior to its binding) the conformation of the target may represent an active or inactive intermediate state, which is ‘locked’ after the ligand has bound. Another possibility is the interference of ligand binding on signal transduction pathways at sites of protein-protein interaction. Besides soluble proteins, other target classes such as proteins embedded in membranes, nucleic acids (DNA/RNA) or even proteoglycans can be addressed by affinity selection based HTS (Figure 1). Most of these targets would have to be excluded from HTS if only functional assays were available. In this article we will review HTS technologies with small molecule libraries based on affinity-selection of chemical binders.
Labeled methods
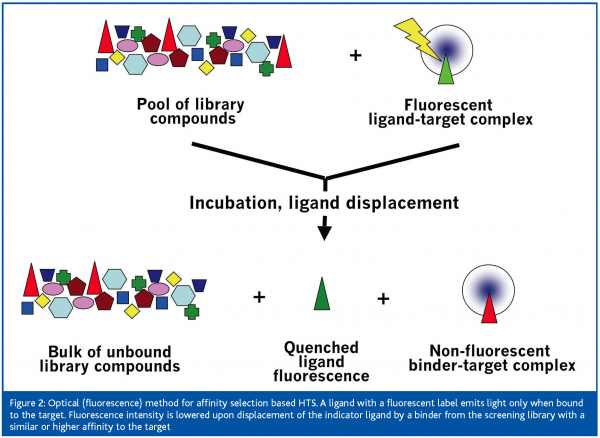
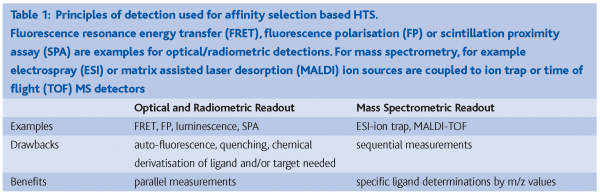
Classical biochemical HTS technologies with miniaturised functional assays, e.g. in 1536-well format, essentially use optical or radiometric detection. Under routine conditions, technologies such as fluorescence resonance energy transfer (FRET), fluorescence polarisation (FP) or luminescence are successfully used to analyse >40,000 compounds per day due to a parallel readout. Optical detection can also be applied to affinity selection based HTS. Such a method would measure the displacement of a known ligand by compounds from the screening library with higher affinity to the target, which translates into an optical signal: for example the light emission of a fluorescent ligand is quenched upon ligand displacement by a compound from the screening library (Figure 2). An alternative to a fluorescent or luminescent readout is the use of radiometric compounds. Scintillation proximity assay (SPA)2 is an example. When applying this technology to affinity selection based HTS, a radio-labeled ligand is non-covalently bound to the immobilised acceptor molecule on a scintillant-containing bead and will induce light emission as long as it is not displaced by a compound from the screening library with similar or higher affinity to the target.
A drawback of all labeled HTS technologies is that they request chemical derivatisation of either the target or the ligand with a fluorometric or radiometric label. This is cost intensive and can alter the target with respect to its biological activity, or the ligand with regard to its specificity and affinity to the target. Another inherent weakness of all HTS technologies with fluorescence readout is their susceptibility to artifacts caused by auto-fluorescence of many library compounds or by non-specific fluorescence quenching of compounds. Whenever these potential drawbacks can be avoided or controlled, labeled HTS methods are a powerful means to perform not only affinity-selection based high-throughput screening, but also HTS with functional assays. Table 1 compiles several detection modes used in affinity selection based HTS.
Label-free methods
Label-free methods are technologies where neither the compounds or the target biomolecules are modified by a fluorometric or radiometric label. Mass spectrometry (MS) is an appropriate tool to identify binder molecules or the target-binder complex by their mass to charge (m/z) ratios.
Direct ligand detection by MS
Non-covalent binder-target complexes can be directly determined following soft ionisation either with matrix assisted laser desorption ionisation (MALDI) or electrospray ionisation (ESI) devices coupled to a mass spectrometry detector3. Common to both approaches is the direct measurement of the shift in molecular weight (m/z) of the charge isomers of the ligand-free target upon formation of the ligand-target complex.
Whereas ESI mediated determinations of protein-binder complexes are limited to protein masses <50,000 Da, MALDI coupled to a time-of-flight (TOF) MS detector enables the detection of biomolecules of >300,000 Da molecular mass. Investigations of non-covalent binder-target complexes by MALDI appear, however, to be limited to those with higher binding energies4,5.
Indirect ligand detection
When using an indirect detection of binder molecules, the non-covalent binder is separated from the target prior to the binder identification. Electrospray ionisation (ESI) rather than atmospheric pressure chemical ionisation (APCI) is used for binder identification by its m/z value in the mass spectrometry device. Compared to parallel detection with optical (e.g. fluorescence) methods, MS detection in HTS is relatively slow due to its sequential readout. There is a necessity to use pools of the library compounds when analysing several 10,000 compounds per day with MS. Both the pool size and compound composition must be carefully addressed, because chemical cross-reactivity and the solubility of the mixed compounds set limits to the pool complexity. Pool sizes of up to 2,500 compounds per well have been reported in the literature11. With another affinity selection HTS technology8,9, 400 compounds per pool are not exceeded as substances with higher molecular masses than any of the pooled compounds have been detected when using pools with >500 compounds per well. The appearance of new substances with higher molecular weight probably reflects chemical cross reactivity. Attention must also be paid to potent solvents, such as DMSO. These are favourably used to dissolve the heterogenic collection of the chemical compounds, but the potential harm on biological integrity of the target protein needs to be balanced with the dissolution power. DMSO concentrations <5 per cent seem to be tolerated by most of the proteins.
Heterogeneous and homogenous formats
When an indirect method is used for the screening of binder molecules from a compound pool, affinity selection must distinguish between target-bound and unbound compounds and must deliver a reliable format for binder identification.
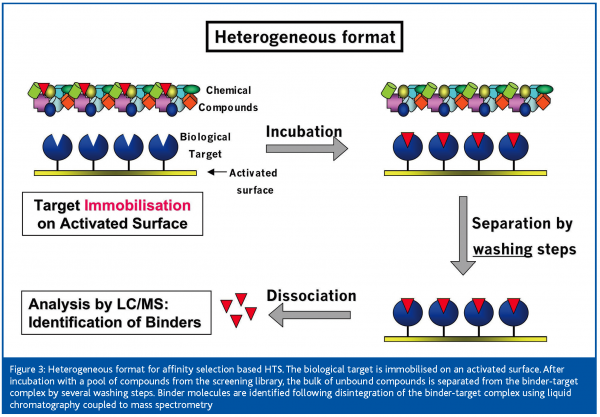
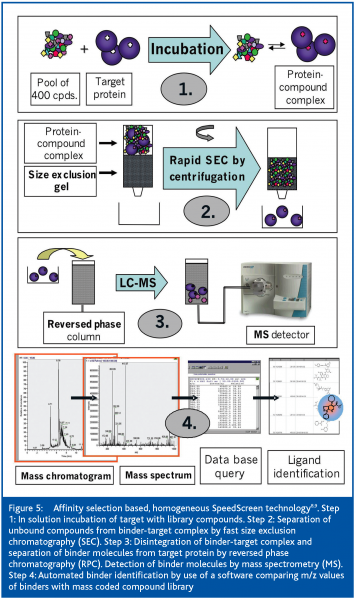
With the heterogeneous format [Figure 3], the target is immobilised on an activated surface. After incubation with a pool of compounds, unbound substances are removed from the target-binder complex in several washing steps. The continuous depletion of the binder concentration by the washing solution leads to substantial losses of non-covalently bound ligands with low target affinity. Only for a stronger binder will a sufficiently high number of molecules remain bound to the target upon repeated washing to be measurable by, for example, a sensitive MS detector. Therefore, affinity selection based HTS using a heterogeneous method is selecting for high-affinity binders. A screening method called frontal affinity chromatography-mass spectrometry (FAC-MS) was published recently7. Immobilised target proteins are used to determine affinity ranking and Kd values of ligands by a chromatographic process. For this purpose, a compound mixture is continuously infused over the target, which is immobilised on the solid support (beads) in the column and the effluent is monitored on-line by MS. Based on their affinity to the target, the compounds are more or less efficiently retained by the target. The retention time of a binder’s break-through in the column effluent can be set in relation to two reference ligands with a known high and low affinity to the target, in order to estimate affinity ranking or affinity constants of binders. Screening of pools of up to 200 compounds per sample has been claimed, which requests powerful software for data evaluation. Despite the pooled screening approach, FAC-MS appears to be better suited for secondary assays than for primary screening in the HTS process.
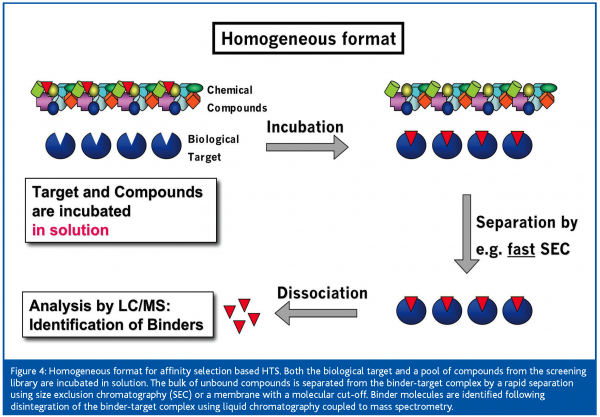
In the homogeneous format [Figure 4] both target and compounds are combined in solution. When using compound pools for incubation with the target, a fast one-step separation is crucial to remove the bulk of unbound compounds before detection of binder molecules. For the separation of target-bound from unbound compounds, essentially size exclusion chromatography8,9,10 (SEC) or membrane filtration with an appropriate molecular weight cut-off6 have been used. Both approaches focus on a rapid and effective separation of the target-binder complex from unbound compounds in order to minimise losses in target-bound ligands. During SEC, dissociation of bound molecules from the target-binder complex, due to the continuous depletion of binder molecules along the way of the binder-target complex through the size exclusion gel, must be minimised. Therefore, the separation step must be fast and conducted at low temperature in order to prevent the loss of too many binder molecules required for MS detection.
When using a competitor ligand with a known affinity constant to displace a target-bound compound from the screening library, affinity ranking of binders is possible10,11. In equilibrium affinity selection-mass spectrometry11, the target is first incubated with the library compounds before separation of the target-binder complex from unbound substances, similar to the procedure used in another homogeneous method8,9. Compounds with a constant concentration and a variety of affinities to the target are then submitted to this procedure in the presence of serially increasing concentrations of a known competitor. MS measurements of binders and competitor allow the determination of the affinity constants (Kd) of each component.
An alternative to the detection of bound ligands together with the target biomolecules was recently described12. In this method, low affinity binders were incubated at equally molar amounts with the target and analysed in fractions collected after the high molecular weight substances, i.e. the target or the target-binder complex, had eluted from the SEC gel. A strong binder to the target would not be detected in the low molecular weight fraction of the SEC eluate. With this medium- rather than high-throughput screening method, affinity ranking of binder molecules was demonstrated when applying small mixtures of protein ligands to SEC separation and sequentially excluding the molecule which was not detected in the low molecular weight fraction from the next incubation with the target protein.
Multiplexing
Multiplexing the SEC separation step is mandatory to match high-throughput screening conditions. The ALIS method uses an array of SEC columns13 for this purpose. Another technology, termed SpeedScreen8,9, is based on a 96-well microtiter plate format. Upon short centrifugation, 96 samples can be chromatographed at the same time in one short SEC step with an estimated effective separation time of less than ten seconds. SpeedScreen was developed especially for high-throughput screening (HTS) of orphan and non-tractable targets. Following in-solution affinity selection with compound pools, binder molecules are separated from unbound compounds by size-exclusion chromatography (SEC). Disintegration of the binder-target complex is achieved by microbore-liquid-chromatography and binder identification by electrospray-ionisation and mass spectrometry measurements (µLC/ESI-MS). Details of the SpeedScreen methodology were recently published in detail8,9 and are depicted in Figure 5.
As with all homogeneous methods using compound pools for screening, an efficient separation of target-bound from unbound compounds is crucial. In order to keep a sufficiently high number of target-bound ligands that match the sensitivity of the MS detector, both a sufficiently high target concentration and a short time for ligand separation from non-bound compounds are essential. Without these precautions, binders with a high koff rate are most likely lost during a slow separation procedure. When conducting the separation step sufficiently fast, however, even moderately strong binders can be detected. This appears to be an advantage over the heterogeneous methods using washing steps for binder separation. Attention should also be drawn to the efficient disintegration of the binder-target complex prior to binder identification. Chromatography of the complex with an organic solvent gradient at an elevated temperature (e.g. 60°C) was successfully used to dissociate most of the high-affinity ligands from the target protein. As MS detection parameters cannot be optimised for each of the hundreds of thousands of library compounds, a generic method must be applied with sensitive MS detection to scan the mass range of the compounds used for screening in each sample. Ion trap MS detectors have an advantage over, for example, triple quadrupole detectors that their sensitivity in full scan mode is comparable to that in single ion monitoring. Linear ion traps are even close to the sensitivity of triple quadrupoles. However, both types of MS detectors lack high mass resolution compared to a time of flight (TOF) or Fourier transform ion cyclotron resonance (FTICR) MS detector. This low precision in mass determinations of the ion traps is a drawback when screening compound pools in single stage MS (MS1) mode. The more specific measurements of compound fragments (daughter ions) in MS-MS (MS2) mode are at least difficult when screening pools of several hundreds of compounds per sample, as this procedure would require the generation of huge libraries with daughter ions of all library compounds used for screening. With single stage MS, however, the appearance of isobars, i.e. ions with the same m/z value but related to compounds with a different chemical structure, must be anticipated. Ideally, the pools would be mixed in a way that ensures the appearance of isobars is avoided. This is not an easy goal when compounds from libraries with more than one million distinct entities are pooled.
Data evaluation
There is a huge set of analytical data when using MS to identify binder molecules with a particular target, which requests time consuming data evaluation. Software enabling automated data evaluation is helpful to avoid the tedious manual efforts. Software for this purpose is often written by screeners themselves, who know what features the software should have in order to enable desired data handling. In a few cases, such software is co-developed by a company conducting HTS on a routine basis and a company providing IT expertise. Refer to the Web sites of such software companies in the public domain, for more information.
Binder characterisation
In contrast to hit lists from a functional HTS containing, for example, IC50 data, delivery of affinity-selection based HTS is often restricted to lists of binder molecules with no further characterisation. Secondary biochemical profiling of the binders, by means of IC50 or Ki determinations, is performed in case the target has an enzymatic or a receptor function. With cellular assays, the influence of a binder on a receptor (e.g. GPCR) mediated signal transduction can be measured, for example, by the modification of intracellular c-AMP or calcium levels, using equorin or other Ca++-dependent fluorescing indicators for the latter. Another cellular system may take advantage of a reporter gene assay to visualise the effect of a binder molecule on a target which is part of a particular signaling pathway and leads to a specific gene expression linked to the expression of a reporter gene.
For binders of targets, however, which do not have an enzymatic function or where the target’s function is unknown, determination of an affinity constant is often the only possibility for binder characterisation. Some technologies, such as nuclear magnetic resonance (NMR), surface plasmon resonance (SPR), microcalorimetry, acoustic profiling or the above mentioned FAC-MS are potential options to characterise binder molecules. Although all these technologies have the potential for affinity selection screening, they cannot be attributed to high-throughput screening in a stringent sense due to their limited throughput.
Conclusions
Affinity selection based HTS is a complementary screening technology to functional HTS. It widens the scope of HTS to orphan and non-tractable targets, which are not amenable to functional HTS. Both heterogeneous and homogeneous formats have been successfully used. Optical readouts are feasible with affinity selection based HTS, but highly sensitive mass spectrometry enables the use of label-free methods. In case emphasis is essentially put on high throughput, mainly binder information can be delivered by affinity selection based HTS. With known affinity-characterised ligands, affinity ranking or even Kd determinations of binder molecules are feasible, but at the expense of a decreased throughput. An alternative approach is binder characterisation from the affinity selection based primary HTS by a subsequent biochemical or cellular assay to generate IC50 or Ki data.
Acknowledgement
The authors are very grateful to all colleagues at NIBR (Novartis Institute of Biomedical Research), Department of Discovery Technologies, who have contributed to the successful implementation and application of the affinity selection based screening technologies, in particular René Amstutz, Manfred Auer, Pascal Bernet, Jutta Blank, Ireos Filipuzzi, Michael Forstner, Felix Freuler, Peter Fuerst, Fraser Glickman, Martin Klumpp, Lukas Leder, Natalie Lehmann, Ingo Muckenschnabel, Reto Naef, Johannes Ottl, Thomas Stettler and Claudia Textor. We fully appreciate their continuous support, encouragement and enthusiasm for novel technologies in drug discovery.






References
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG et al. 2001 The sequence of the human genome. Science 291, 1304-1354
- Bosworth, Towers P. 1989 Scintillation proximity assay. Nature 341, 167-168
- Pramanik BN, Bartner PL, Mirza UA, Liu Y-H & Ganguly AK. J. 1998. Electrospray Ionization Mass Spectrometry for the Study of Non-covalent Complexes: an Emerging Technology. Mass Spectrom. 33, 911-920
- Karas M, Bachmann D, Bahr U & Hillenkamp F. 1987. Matrix- Assisted Ultraviolet-Laser Desorption of Nonvolatile Compounds. Int. J. Mass Spectrom. Ion Processes 78, 53-68
- Hillenkamp F, Karas M, Beavis RC & Chait BT. 1991. Matrix-Assisted Laser Desorption Ionization Mass-Spectrometry of Biopolymers. Anal. Chem. 63, A1193-A1202.
- Deharo E, Garcia RN, Oporto P, Gimenez A, Sauvain M et al. 2002. A non-radiolabelled ferriprotoporphyrin IX biomineralisation inhibition test for the high throughput screening of antimalarial compounds. Exp. Parasitol. 100, 252-256
- Slon-Usakiewicz JJ, Ng W, Dai J-R, Pasternal A & Redden PR. 2005. Frontal affinity chromatography with MS detection (FAC-MS) in drug discovery. Drug Discov. Today 10, 409-416
- Muckenschnabel I, Falchetto R, Mayr LM, Filipuzzi I. 2004. SpeedScreen: label-free, liquid-chromatography-mass spectrometry-based high-throughput screening for the discovery of orphan protein ligands. Anal. Biochem. 324, 241-249
- Zehender H, Le Goff F, Lehmann N, Filipuzzi I & Mayr LM. 2004. SpeedScreen: The “Missing Link” between Genomics and Lead Discovery. J. Biomol. Screen. 9, 498-505
- Annis DA, Nazef N, Chuang Ch-Ch, Porter Scott M & Nash HM. 2004. A General Technique to Rank Protein-Ligand Binding Affinities and Determine Allosteric versus Direct Binding Site Competition in Compound Mixtures. J. Am. Chem. Soc. 126, 15495-15503
- Whitehurst CE, Nazef N, Annis DA, Hou Y, Murphy DM, Spacciapoli P, Yao Z et al. 2006. Discovery and Characterization of Orthosteric and Allosteric Muscarinic M2 Acetylcholine Receptor Ligands by Affinity Selection-Mass Spectrometry. J. Biomol. Screen. 11, 194-207
- Wabnitz P A, Loo JA. 2002. Drug screening of pharmaceutical discovery compounds by micro-size exclusion chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 16, 85-91
- Lenz GR, Nash HM, Jindal S. 2000. Chemical Ligands, Genomics and Drug Discovery. Drug Discovery Today, 5, 145-156




