Detecting and eradicating mycoplasmas in ATMP cell lines
Posted: 14 June 2022 | Hannah Balfour (European Pharmaceutical Review) | No comments yet
Here, EPR summarises the key points of Tim Sandle’s recent review of mycoplasma risks, detection and eradication techniques for cell cultures.

Mycoplasmas are a concern for cell culture viability – and difficult to detect in the advanced therapy medicinal products (ATMPs) that cell cultures produce. The presence of mycoplasmas, ie, bacteria without cell walls, in cell therapies could cause side effects in the immunocompromised patients they may be used to treat.
Within a cell culture, mycoplasmas alter cell physiology, impacting the integrity of cellular components critical for the ATMP product, as well as lead to the loss of cell lines. Though rates of contamination vary, a contaminated eukaryotic cell line can contain 106-108 organisms/ml.
According to Tim Sandle of Bio Products Laboratory, mycoplasma contamination is the “foremost concern” for stakeholders developing and using cell lines, not because it happens more frequently than contamination with other bacteria and fungi, but because it is less readily detectable. Additionally, mycoplasmas can spread vastly among cell cultures.
To prevent or control this type of contamination, manufacturers must consider their potential sources (personnel, materials, consumables, equipment, media and reagents) and take steps to minimise the risk of contamination from the environment or cross-contamination from other products.
How are mycoplasmas detected?
Mycoplasmas cannot be detected with the naked eye, instead culture-based, DNA staining and polymerase chain reaction (PCR) methods are used.
Culture-based methods are the standard; however, are limited by the long growth times required and the presence of uncultivatable species. Additionally, complex media is required to support the growth of mycoplasmas, resulting in variability in manufacture that requires additional QC.
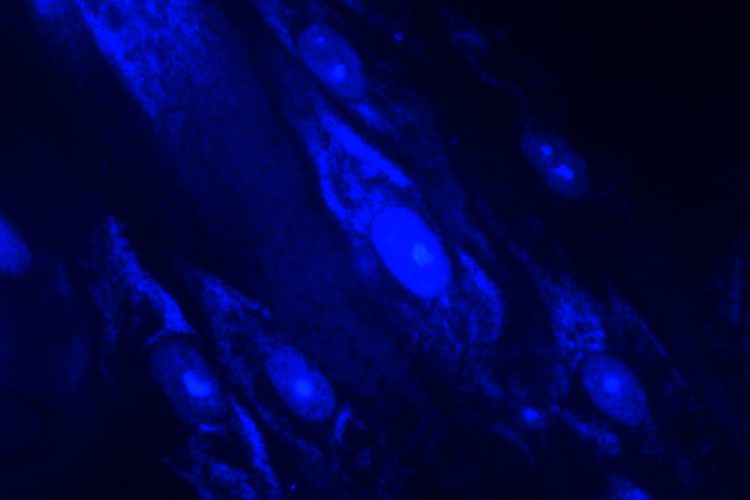
Staining can be used as an adjunct to culture-based methods, leveraging DNA fluorochrome staining (such as the Hoechst stain) and ultraviolet light to visualise cells and contamination with fluorescence microscopy. Samples infected with mycoplasmas are seen as fluorescing nuclei plus extra-nuclear fluorescence of mycoplasma DNA (small cocci or filaments).
While culture-based methods remain important, faster diagnosis can be obtained through PCR – and the low signal of staining methods make PCR the detection method of choice, wrote Sandle.
The PCR test is based on the detection of 16S and 23S rRNA molecules of the most common species of mycoplasma contaminating cell cultures. PCR techniques provide sensitive, specific and rapid diagnosis of mycoplasmal infection, noted Sandle, with rapid tests enabling detection of one or two hundred mycoplasma species within around two hours. PCR can be strengthened through enrichment, where a growth enrichment step is included prior to the nucleic acid test to delineate viable organisms from non-viable organisms and residual environmental sources. Without enrichment, the methods do not allow for accurate discrimination between viable and non-viable contaminants, which might lead to false-positive results.
While PCR testing is used more often for the detection of mycoplasmas, there is no general approval for PCR-based mycoplasma testing by the regulating agencies. Therefore, when developing a PCR method, wrote Sandle, a comparability study should be undertaken with the culture method.
What eradication techniques are there?
Attempts can be made to remove mycoplasmas from contaminated cell cultures; however, the processes are complex. Filtration and the application of ultraviolet light are the most common, additionally certain antibiotics can be used. With filtration, mycoplasma clearance requires 0.1μm rated sterilising filters, as the bacteria are small enough to penetrate 0.22μm grade filters.
If eradication is unfeasible, the culture should be disposed of.
The review was published in the Journal of Validation Technology (JVT).
Related topics
Analytical techniques, Biologics, Bioproduction, Drug Manufacturing, Drug Safety, Microbial Detection, Microbiology, QA/QC