Established and new mass spectrometry applications to drug discovery and development
Posted: 18 February 2021 | Dr Ioannis Papayannopoulos | No comments yet
Mass spectrometry has numerous applications in pharmaceutical research, from initial discovery through to characterisation and quality control. In this article, Dr Ioannis Papayannopoulos details the role of mass spectrometry in pharma R&D by highlighting established and novel applications of the technique in drug discovery and development.


For over half a century mass spectrometry has been used in the pharmaceutical and biotechnology industries (hereafter collectively referred to as biopharmaceutical); initially in discovery research and subsequently in the development, characterisation and quality control of pharmaceuticals, both for small molecule drugs and biologics. As instrument and software improvements are made and new technologies are developed, mass spectrometry has continued to expand its role in drug discovery and development and make inroads in new areas of pharmaceutical and biopharmaceutical R&D.
Established mass spectrometry applications
Instrumentation
Gas chromatography coupled to mass spectrometry (GC-MS)1 was the earliest “hyphenated” mass spectrometric method used extensively in commercial enterprises; it is applicable to molecules that are volatile and relatively stable at the higher temperatures required for gas chromatographic separations. GC-MS applications span a variety of areas of research and development (environmental monitoring, sampling and analysis; characterisation and quantitation of volatile metabolites from biological processes such as respiration or fermentation; monitoring production processes) in many diverse industries (fine chemicals, foods and beverages, plastics, paints, electronics). However, most drugs that are being developed and marketed comprise polar, non-volatile, thermally fragile molecules, thus liquid chromatography coupled to mass spectrometry (LC-MS), the latter typically equipped with electrospray ionisation (ESI), is the most important and widely used mass spectrometric method in the biopharmaceutical industry. Matrix-assisted laser desorption ionisation (MALDI) mass spectrometry, usually implemented on a time-of-flight (TOF) mass analyser, is not nearly as widespread but remains important for certain applications.
Applications to discovery research
In discovery, the identification of useful drug targets through the deconvolution of biological pathways has been greatly facilitated by mass spectrometry. The field of proteomics – the identification and quantitation of differentially expressed proteins in normal versus perturbed (disease and drug-treated) biological systems – has found numerous beneficial applications. It has enabled faster and more robust discovery and validation of drug targets, earlier identification of potential off-target activities of drug candidates and the discovery and validation of biomarkers associated with a disease state, progression and treatment, which is important in assisting with disease diagnosis and facilitating monitoring of treatment.2
Mass spectrometry has long been used to characterise drug candidates, confirm that the desired molecule has, indeed, been made and identify, with very high sensitivity and accuracy, impurities – the presence of which may obfuscate attempts to derive structure-activity relationships.3 Fast analyses with methods amenable to multiplexing have enabled the rapid identification and characterisation of potential hits in high‑throughput screening of large compound libraries.4
Applications to development, drug metabolism and pharmacokinetics
Moving into drug development, mass spectrometry serves as a helpful guide during formulation development, especially in assessing the stability of potential drugs in different formulations.5 Further along the drug development process, mass spectrometry has been an essential method for understanding drug metabolism and the pharmacokinetic (DMPK) properties of drug candidates, in both animal models and humans.6 This involves extraction of the drug of interest and its potential metabolites from plasma or other tissues and the analysis and quantitation of the various molecules present to gather information on the in vivo stability, distribution and metabolite profile of a drug. Pharmaceutical companies have entire departments equipped with numerous mass spectrometers (mostly triple quadrupole instruments), dedicated to these analyses; many contract research organisations (CROs) offer such services to the pharmaceutical industry.
While the use of mass spectrometry for DMPK analyses of small molecule drugs spans half a century, its application to similar analyses of biological drugs (proteins, antibodies, nucleotides) is a more recent development. The reason for the later entry of mass spectrometry to biologics DMPK is related to the instruments’ capabilities in identifying and accurately quantitating large molecules, which are often difficult to separate from other proteins present in plasma or tissues and difficult to ionise efficiently. Great improvements in mass spectrometer resolution, mass accuracy and sensitivity, coupled with significant improvements in liquid chromatography systems and columns, have enabled direct measurement of intact proteins in serum and the detection and quantitation of peptides, obtained by proteolytic digestion, that are unique to the protein drug of interest.7 The use of analogues synthesised with stable isotopes (13C, 15N, 18O) and spiked into the sample in well‑defined concentrations (“isotopic dilution”), long used for accurate quantitation of small molecule drugs and metabolites in biological fluids, is also applied to the quantitation of proteolytic peptides from protein drugs.8
Applications to drug product characterisation and release
For any drug candidate for which regulatory permission is sought for its use in clinical investigations, a whole array of testing is required to demonstrate that it is well characterised, its structure is elucidated in detail, its impurity profile is known and quantified within defined ranges, its stability is monitored and its manufacturing process is controlled and reproducible. The stringency of these requirements increases as a drug progresses through the different phases of clinical trials and is eventually licensed for commercial release. Although it is by no means the only analytical method employed for characterisation, mass spectrometry is a critical one, equally useful for small molecules and biologics.9
Typically, an informative subset of the analytical methods used for characterisation is applied to the release of clinical and commercial drugs. Batch release testing is carried out under well‑defined guidelines,10 generally agreed upon and enforced worldwide by the various regulatory agencies (eg, the US Food and Drug Administration [FDA] in the US, the European Medicines Agency [EMA] in the EU, and the Medicines and Healthcare products Regulatory Agency [MHRA] in the UK). While the application of mass spectrometry to release testing of small molecule drugs has been used for some time, it has not been generally employed in batch release testing of biologics (despite playing a critical role in product characterisation). This has been due to difficulties in validating the preparation of biologics for analysis, as well as the instruments themselves. Simply put, preparing samples and operating the instruments to generate good mass spectrometric data from large molecules with proper adherence to good manufacturing practices (GMP) guidelines on a routine basis has been far more involved, cumbersome and complicated than it has been for small molecule drugs.
New mass spectrometry applications
Multi-attribute method for protein drugs
In the past few years, several groups have demonstrated that it is possible to replace some of the release testing methods typically used for protein pharmaceuticals with a single mass spectrometry-based multi-attribute method (MAM),11,12 as several attributes, each measured individually by a different release method, can be identified and quantified by a single MAM, as shown in Table 1.
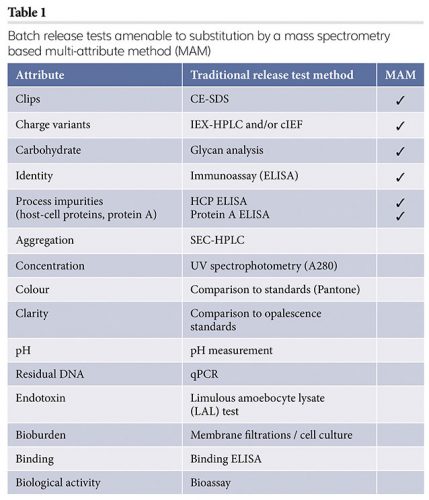

Adoption of MAM by the biopharmaceutical industry has been gaining momentum, driven by improvements in the capabilities of chromatography and mass spectrometry instruments and data acquisition and processing software. For a MAM to perform well, new species generated from changes to a molecule, due to unexpected variations in the manufacturing process or to handling and storage, must be separated chromatographically, quantitated accurately and the resulting mass spectra measured with great sensitivity, mass resolution and mass accuracy. Additionally, the front-end sample preparation must be streamlined and automated for consistency and reproducibility. The major chromatography and mass spectrometry vendors have been working closely with early adopters of this approach in the biopharmaceutical industry to implement, improve and refine MAM hardware and software applications.
Tissue imaging
Tissue imaging involves scanning a tissue sample with a laser, which has been introduced into the ion source of a MALDI-TOF mass spectrometer, and recording signals from the ionised molecules present. It is proving to be very useful in clinical settings for obtaining in a matter of hours biomarker information indicative of disease and disease progression, faster than immunohistochemistry and without requiring specific antibodies.13 In drug discovery and development, tissue imaging can generate information on bioavailability, in situ metabolism and the spatial distribution in different tissues and organs of small molecule drugs and of toxicity biomarkers.14 This method has the distinct advantage of being applicable at earlier stages of drug discovery when hundreds of drug candidates can be screened in tissue samples from animal models instead of relatively few lead molecules at a later stage in development. This is because tissue imaging with mass spectrometry requires no antibodies for each individual drug candidate; generating such antibodies for immunostaining takes time and is expensive. The coupling of MALDI ionisation to high resolution and high mass accuracy commercial TOF and trap mass analysers, along with the development of sophisticated software, has made this approach routinely available to pharmaceutical research and development analytical laboratories. However, thus far, protein drugs have not been amenable to tissue imaging due to their high mass and lower ionisation efficiency compared to small molecules.
Hydrogen-deuterium exchange, protein footprinting and crosslinking
Hydrogen-deuterium exchange,15 protein footprinting16 and protein crosslinking17 each provide insights into protein folding and binding, and to the interactions of proteins with other molecules. Hydrogen-deuterium exchange is reversible and requires rapid enzymatic digestion and fast chromatography at lower temperatures to preserve the deuterium substitutions of exchangeable hydrogens. Protein footprinting, which involves oxidation of solvent‑accessible parts of a protein by hydroxyl radicals, and crosslinking, in which covalent bonds are formed either intramolecularly, thus “freezing” a particular folded configuration of a protein molecule, or intermolecularly, covalently linking two or more molecules, are more permanent. Specialised equipment to generate the hydroxyl radicals for protein footprinting and photoactivated linkers to join molecules interacting at specific distances are required, in addition to specialised software to process the mass spectrometric data. These methods are being used in the biopharmaceutical industry to characterise the interaction of protein drugs with small molecules of other proteins (eg, epitope mapping of antibodies), understand protein folding and provide an orthogonal method to size exclusion chromatography in assessing the extent of aggregation of protein drugs.
Metabolomics
In a sense, metabolomics is analogous to proteomics; instead of proteins, small molecules and metabolites are identified and quantified. The change in the small molecule complement of a system (cell, tissue, organ) owing to disease, drug treatment or other perturbations, measured in a quantitative fashion using mass spectrometry, can provide insights on disease progression, efficacy of a therapeutic regimen or toxicity from administration of a drug. Metabolomics remains primarily a research activity; however, there is great potential for its application to drug development by comprehensively monitoring the effects of distribution, efficacy and toxicity of drug candidates.18
Native mass spectrometry of proteins and protein complexes
Native mass spectrometric analysis of large protein complexes preserves non-covalent interactions, which are typically disrupted by most methods of sample introduction (eg, reversed phase HPLC) or ionisation.19 Thus, the mass and stoichiometry of multiprotein complexes can be measured and by selective disruption of the protein complex in the mass spectrometer a three-dimensional (3D) image can be generated, revealing spatial arrangements of the proteins present. Applied to structural biology, native mass spectrometry has yielded insights into the oligomeric states of proteins, the composition and heterogeneity of protein complexes and interactions between different proteins. In drug discovery and development, native mass spectrometry can be used to screen compound libraries, study amyloid formation and its inhibition and disruption by small molecules or proteins and characterise the interaction of drugs with protein targets.20 An ion mobility interface to a mass spectrometer, providing a mode of separation of gas phase ions based on their shape and which has been used to reduce background noise in small molecule studies,21 offers an additional dimension of separation in the mass spectrometric analyses of protein complexes in their native state.22
Conclusion
Mass spectrometry is a well-established method for the analysis of both small molecule drugs and biologics and it continues to make inroads into all areas of biopharmaceutical research and development. In some cases, such as assessing bioavailability, drug metabolism and pharmacokinetics, where mass spectrometry has been critical for small molecule drug development, this role is being extended into similar analyses of protein pharmaceuticals. In other areas, mass spectrometry-based novel techniques such as metabolomics, tissue imaging and methods for generating information on protein folding and protein-protein interactions, are being applied to drug discovery and development.
Dr Ioannis A Papayannopoulos
Ioannis has been carrying out peptide and protein analytical work for the past several decades, using mass spectrometry, chromatography, spectroscopy and other analytical techniques. He received his undergraduate degree in chemistry from Bowdoin College and his PhD in Organic Chemistry from the Massachusetts Institute of Technology under the supervision of the late professor Klaus Biemann. Over the years Ioannis has alternated between academia, most recently as director of the Proteomics Facility at the Koch Institute for Integrative Cancer Research at MIT and the biopharmaceutical industry in senior scientific and management roles at companies such as Biogen, Covance and AstraZeneca. For the past six years he has been with Celldex Therapeutics working on the analysis and characterisation of antibodies, antibody-drug conjugates and recombinant protein pharmaceuticals.
References
1. Sparkman DO, Penton Z, Kitson FG. Gas Chromatography and Mass Spectrometry: A practical Guide. Academic Press, 2011.
2. Cutler O, Voshol H. “Proteomics in pharmaceutical research and development”, Proteomics Clinical Applications 9, 643-650 (2015).
3. Gillespie TA, Winger BE. “Mass spectrometry for small molecule pharmaceutical product development: A review”, Mass Spectrometry Reviews 30, 479–490 (2011)
4. Rohman M, Wingfield J. “High-throughput screening using mass spectrometry within drug discovery”, Methods in Molecular Biology 1439, 47-63 (2016).
5. Chutvirasakul B, et al. “Development and applications of liquid chromatography-mass spectrometry for simultaneous analysis of anti-malarial drugs in pharmaceutical formulations”, Journal of Pharmaceutical and Biomedical Analysis 195 (2021), in press.
6. Cuyckens F. “Mass spectrometry in drug metabolism and pharmacokinetics: Current trends and future perspectives”, Rapid Communications in Mass Spectrometry 33, 90-95 (2019).
7. Gao Y, et al. “Liquid chromatography-mass spectrometry method for the quantification of an anti-sclerostin monoclonal antibody in cynomolgus monkey serum”, Journal of Pharmaceutical Analysis (2021), in press.
8. Heudi O. “Towards Absolute Quantification of Therapeutic Monoclonal Antibody in Serum by LC−MS/MS Using Isotope-Labeled Antibody Standard and Protein Cleavage Isotope Dilution Mass Spectrometry”, Analytical Chemistry 80, 4200–4207 (2008).
9. Deepali R, et al. “The role of mass spectrometry in the characterization of biologic protein products”, Expert Review of Proteomics 15, 431-449 (2018).
10. CFR – Code of Federal Regulations Title 21 [Internet]. Accessdata.fda.gov. 2021 [cited 22 January 2021]. Available from: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfCFR/CFRSearch.cfm?fr=211.165
11. Rogstad S, et al. “Multi-attribute method for quality control of therapeutic proteins”, Analytical Chemistry 91, 14170-14177 (2019).
12. Ren D. “Advancing mass spectrometry technology in cGMP environments”, Trends in Biotechnology (Special Issue: Therapeutic Biomanufacturing) 38, 1051-1053 (2020).
13. Basu SS, et al. “Rapid MALDI mass spectrometry imaging for surgical pathology”, NPJ Precision Onclology 17 (2019).
14. Schulz S. “Advanced MALDI mass spectrometry imaging in pharmaceutical research and drug development”, Current Opinion in Biotechnology 55, 51-59 (2019).
15. Deng B, et al. “Hydrogen deuterium exchange mass spectrometry in biopharmaceutical discovery and development – A review”, Analytica Chimica Acta 940, 8-20 (2016).
16. Wang L, Chance MR. “Protein footprinting comes of age: Mass spectrometry for biophysical structure assessment”, Molecular & Cellular Proteomics 16, 706-716 (2017).
17. Sinz A. “Cross-linking/mass spectrometry for studying protein structures and protein-protein interactions: Where are we now and where should we go from here?”, Angewandte Chemie (International Edition in English) 57, 6390-6396 (2018).
18. Puchades-Carrasco L, Pineda-Lucena A. “Metabolomics in pharmaceutical research and development”, Current Opinion in Biotechnology 35, 73-77 (2015).
19. Richards AL, et al. “Mass spectrometry‐based protein–protein interaction networks for the study of human diseases”, Molecular Systems Biology 17, 1-18 (2021).
20. Robinson CV, et al. “Using native mass spectrometry to inform drug discovery”, Drug Target Review 2 (2017).
21. Lapthorn C, et al. “Ion mobility spectrometry‐mass spectrometry (IMS‐MS) of small molecules: Separating and assigning structures to ions”, Mass Spectrometry Reviews 32, 47-71 (2013).
22. Stojko J, et al. “Ion mobility coupled to native mass spectrometry as a relevant tool to investigate extremely small ligand-induced conformational changes”, Analyst 140, 7234-7245 (2015).
Issue
Related topics
Analytical techniques, Biopharmaceuticals, Drug Development, Drug Discovery, Mass Spectrometry, metabollic imaging, Research & Development (R&D), Technology