A new small molecule drug discovery and development platform for HLA-associated autoimmune diseases
Posted: 23 April 2021 | Robert B Perni, Ryan J Schutte | No comments yet
In the arsenal of approved small molecule drugs, effective, genetically-targeted therapies for the treatment of autoimmune diseases are conspicuously scarce. ImmunoMolecular Therapeutics (IM Therapeutics) has developed a paradigm for the discovery and development of new molecular entities as a broad platform for blocking genetic targets that confer risk of disease development and are key to triggering autoimmune responses. Ryan J Schutte and Robert B Perni explain more.


IM Therapeutics is developing first-in-class drugs for type 1 diabetes and coeliac disease, as lead programmes within the autoimmune spectrum, by targeting specific human leukocyte antigen (HLA) variants. The discovery platform combines state-of-the-art in silico molecular modelling and structure-based drug design with in vitro cell-based assays to develop small molecules to selectively block autoimmunity. IM Therapeutics’ lead drug candidate IMT-002 is currently in late Phase I clinical trials to study its potential for the treatment of recent-onset type 1 diabetes – the first study in which patients are preselected for an HLA variant that is a high-risk factor for diabetes development.
Introduction
Preventing presentation of autoantigens by specific human leukocyte antigen (HLA) variants with small molecules is not an approach that has received significant industry attention as a mode of therapeutic intervention. IM Therapeutics has developed a discovery platform that employs computational approaches to examine the docking of chemical entities within the antigen-binding cleft of HLA molecules followed by cell-based antigen presentation assays and structure-based drug design to develop novel chemical entities (NCEs). These NCEs are further optimised for inhibiting the binding of antigens associated with autoimmune diseases while maintaining HLA functionality and permitting the binding of antigens originating from pathogens.1,2
In silico screening of large libraries of chemical structures is a sophisticated tool in drug discovery programmes, although it has not replaced the high‑throughput screening (HTS) of physical compounds. While in silico screening provides savings in time and cost compared to HTS, we have observed that the generated hits can be of much higher quality than typical HTS too. Hence these initial hits can potentially be further along the hit-to-lead timeline, shortening hit‑to-lead and consequently lead optimisation phases. It is this shortened timeline that makes the IMT platform valuable and unique in the autoimmune drug discovery space.
IM Therapeutics has combined an in silico modelling and screening paradigm along with in vitro assays that interrogate the interaction between HLA, autoantigens and T cells. The platform is designed to identify, characterise and rank in silico hits and to subsequently evolve NECs through hit-to-lead and lead optimisation. The platform has been validated for targeting insulin and gliadin presentation by two HLA variants, HLA-DQ8 and HLA-DQ2, which are significant mechanisms in the underlying pathogenesis of type 1 diabetes and coeliac disease, respectively.3 IM Therapeutics is extending its discovery efforts to other HLA variants and related autoimmune indications where current treatment options are either limited or lacking.
Underlying biology
HLA proteins are expressed on the cell surface and function by binding self or foreign peptides (antigens) and presenting them to T cells. A T cell carries a receptor (TCR) that allows it to recognise a peptide presented by HLA (an epitope). The interaction of a TCR with the HLA-peptide complex initiates T-cell activation (Figure 1) and begins an immune response. These immune responses mediate the clearance of pathogens (response to a foreign antigen) or, in the case of autoimmune diseases, are directed against an individual’s own tissue (response to a self-antigen).
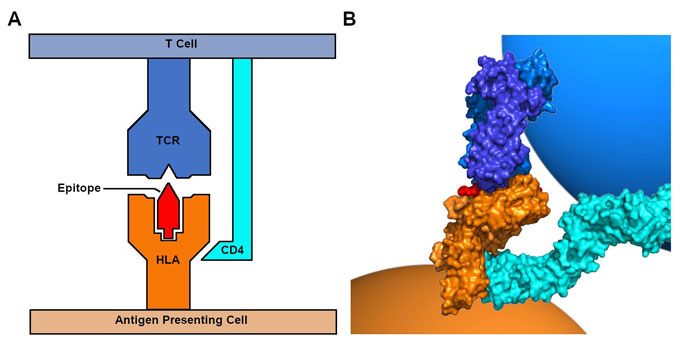

Figure 1: HLAs bind and present antigens to T cells. A. Schematic of the interaction between an antigen‑presenting cell expressing a class II HLA and a CD4+ T cell. B. Image based on the solved crystal structure of the interaction of an HLA-peptide complex with a TCR and CD4 (PDB 3T0E).4 Visualisation colours consistent with labels in A.
The major class I HLAs (HLA-A, B, C) are found on the surface of nucleated cells and present peptides originating from inside the cell to CD8+ T cells. The major class II HLAs (HLA-DQ, DR, DP) are found on professional antigen presenting cells, such as dendrocytes, macrophages and B cells, and present peptides originating from outside the cell to CD4+ T cells. HLAs are highly polymorphic and amino acid differences within the cleft or groove where peptides bind influence the repertoire of peptides an individual HLA molecule can present on the cell surface.
Individual HLA variants confer significant risk for autoimmune diseases.3 HLA-DQ8 is a significant determinant of risk for type 1 diabetes, with ~50‑60 percent of patients being carriers.5 HLA‑DQ2 is carried by ~90 percent of individuals with coeliac disease.6 HLA-DR3 confers risk for multiple autoimmune diseases, including systemic lupus erythematosus and Sjogren’s syndrome.7 HLA-A*29, HLA-B*51 and HLA-C*06:02 are associated with birdshot chorioretinopathy, Behcet’s disease and psoriasis, respectively.3 Studies have indicated that an HLA variant’s preference for antigen binding and presentation contributes to the rise of populations of T cells that recognise self-antigens, have escaped self-tolerance mechanisms and can trigger autoimmunity.3,8 By targeting disease-relevant HLAs, IM Therapeutics’ platform represents a first-in-class personalised therapy approach for autoimmune patients carrying specific disease-associated HLA variants.
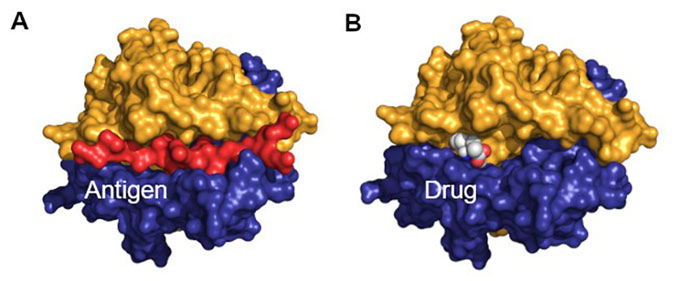

Figure 2: Model of an inhibitor designed to block antigen presentation by an HLA. A. Image of the crystal structure of HLA-DQ8 presenting an insulin peptide (PDB 1JK8).9 The insulin peptide, a self-antigen, is coloured red. B. Model of a small drug, with atoms shown as spheres, designed to bind within the antigen binding cleft and block binding of self-antigens.
The IM Therapeutics in silico platform uses molecular modelling to identify compounds with potential to bind to HLA in order to block antigen presentation (Figure 2).1,2 In silico hits are validated in vitro by cell-based assays using engineered cells that carry a specific HLA variant (eg, HLA‑DQ8) and a second engineered T-cell line that carries a TCR that recognises a specific antigen (eg, Insulin B:13‑23 presented by DQ8).1 This assay system allows assessment of a compound’s ability to disrupt disease-relevant antigen presentation. Additional cell lines carrying other HLAs or TCRs allow for assessment of HLA specificity and if T-cell response to higher affinity pathogen antigens remains intact.
Clinical proof of concept of the approach (IMT-001 and IMT-002)
IM Therapeutics was founded by physician-scientists at the Barbara Davis Center for Diabetes at the University of Colorado. Their foundational work used a crystal structure of HLA-DQ8 as the basis for in silico docking of a library of chemical entities, including FDA-approved drugs.1 In silico triaged compounds were screened for the ability to inhibit presentation of an insulin peptide to an engineered T-cell line by HLA‑DQ8. Several compounds exhibited significant in vitro activity, including the antihypertensive L-α‑methyldopa (Aldomet; IMT‑001, Figure 3). The drug was shown to block antigen presentation in vivo when HLA‑DQ8 transgenic mice were treated orally.1 As L-α-methyldopa has an established safety profile, it was an ideal candidate for a human proof of concept clinical trial (NCT01883804). L-α‑methyldopa blocked HLA-DQ8 presentation of, and reduced the inflammatory T-cell response to, insulin without affecting HLA‑DQ2 or HLA-DR4 antigen presentation.1

Figure 3: Structures of IMT-001 (L-α-methyldopa) and IMT-002 (D-α-methyldopa)
While this result is demonstrative, L-α‑methyldopa is not an ideal candidate because it is an established antihypertensive.10 However, the blood pressure-lowering effect is derived from a metabolite of the drug.11 Drug development efforts around L-α-methyldopa undertaken by IM Therapeutics led to, among other candidates, an enantiomer of Aldomet, D-α-methyldopa (IMT-002), which blocked insulin presentation by HLA-DQ8 but is not metabolised similarly to its L-isomer. Consequently, IMT-002 does not produce measurable antihypertensive activity.12 IM Therapeutics has completed extensive studies of IMT-002 resulting in its clearance as an investigational new drug (IND) and the successful completion of Phase Ia safety assessment. It is currently in a Phase Ib multiple-ascending-dose clinical trial in HLA-DQ8 positive type 1 diabetes patients to study safety and tolerability, pharmacokinetics and activity in inhibiting presentation of self-antigens by HLA-DQ8.
HLA-DQ2 NCE discovery and development – an accelerated platform
Having established proof of concept in targeting HLA-DQ8 for type 1 diabetes, the platform has been extended to other HLA variants, including HLA‑DQ2 to explore therapeutic development in coeliac disease. HLA-DQ2 is carried by 90 percent of coeliac patients and presents antigens originating from gluten.6,13 T cells that recognise these antigens result in the known disease symptoms and mediate small intestine tissue damage.6,14
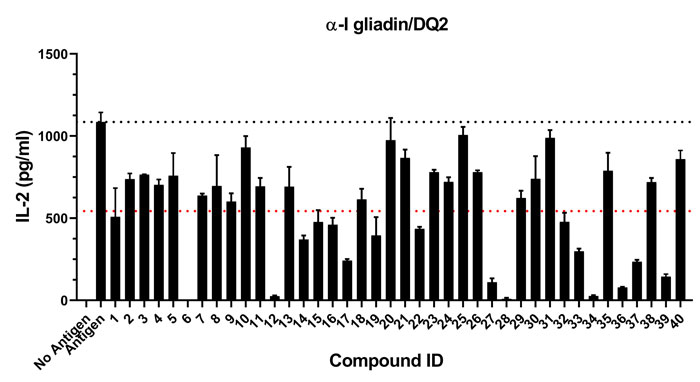

Figure 4: Example data from an HLA-DQ2 in vitro assay of 40 compounds selected by in silico screening. When presented an epitope from α-I gliadin (an antigen originating from gluten), an engineered T-cell line is stimulated and secretes interleukin-2 (IL-2). Compounds that reduce IL-2 secretion, without cytotoxicity or off-target effects (not shown), are characterised to be hits and are further evaluated for pipeline advancement. Dotted lines denote baseline (black) and 50 percent (red) response to antigen.
IM Therapeutics’ platform has advanced to include diverse chemical libraries (>106 molecular entities) covering a significant volume of drug‑like structure-space. In silico top-ranked hits are assessed initially for potential metabolic liabilities, toxicity structural alerts and other standard criteria. Chemical scaffolds are then screened in vitro for blocking antigen presentation (Figure 4) and off-target effects. The platform has yielded diverse structural hits that have been developed into NCE series. Our platform has thus far demonstrated a confirmed hit rate of approximately 10 percent, far higher than typically obtained through HTS. As we enter the lead optimisation phase, we expect to rapidly identify a clinical development candidate to enter IND enabling studies.
Future outlook
IM Therapeutics has demonstrated that the selective disruption of autoantigen presentation by an HLA can be a viable therapeutic strategy for autoimmune disorders. Our evolving discovery platform continues to refine the modelling of HLA‑antigen and HLA-drug interactions in combination with an expanding repertoire of cellular and non-cellular assays. The development pipeline includes additional HLA targets, both class I and class II, with implications in an array of autoimmune disorders.


Ryan is currently a Research Scientist at IM Therapeutics. He holds a PhD in Biomedical Sciences from the University of California, Irvine. His postdoctoral work at the University of Florida focused on modelling HLA‑drug interactions and protein crystallography.


Robert is currently Vice President of R&D at IM Therapeutics. He holds a PhD in organic chemistry from Dartmouth College and has been a medicinal chemist in biopharma for more than 35 years. Dr Perni has extensive experience in drug discovery and development across multiple therapeutics areas including antiviral, antibacterial, oncology, metabolic diseases and autoimmune disorders.
References
- Ostrov DA, Alkanani A, McDaniel KA, et al. Methyldopa blocks MHC class II binding to disease-specific antigens in autoimmune diabetes. J Clin Invest. 2018; 128(5): 1888-1902.
- Ostrov DA, Gottlieb PA, Michels AW. Rationally designed small molecules to prevent type-1 diabetes. Curr Opin Endocrinol Diabetes Obes. 2019; 26(2): 90-95
- Dendrou CA, Peterson J, Rossjohn J, Fugger L. HLA variation and disease. Nat Rev Immunol. 2018; 18(5): 325-339.
- Image of 3T0E (Yin Y, Wang XX, Mariuzza RA. Crystal structure of a complete ternary complex of T-cell receptor, peptide-MHC, and CD4. PNAS. 2012; 109(14): 5405-5410) created with open source PyMOL (The PyMOL Molecular Graphics System, Version 1.8.4 Schrödinger, LLC).
- Erlich H, Valdes AM, Nobel J, et al. Type 1 diabetes genetics consortium. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008; 57(4): 108-92.
- Green PH, Cellier C. Celiac disease. N Engl J Med. 2007; 357(17): 1731-43.
- Gough SC, Simmonds MJ. The HLA Region and Autoimmune Disease: Associations and Mechanisms of Action. Curr Genomics. 2007; 8(7): 453-65.
- Wucherpfennig KW, Sethi D. T cell receptor recognition of self and foreign antigens in the induction of autoimmunity. Semin Immunol. 2011; 23(2): 84-91.
- Image of 1JK8 (Lee KH, Wucherpfennig KW, Wiley DC. Structure of a human insulin peptide-HLA-DQ8 complex and susceptibility to type 1 diabetes. Nat Immunolo. 2001 Jun;2(6): 501-7) created with open source PyMOL (The PyMOL Molecular Graphics System, Version 1.8.4 Schrödinger, LLC).
- van Zwieten PA, Ti-Oolen MJ, Timmermans PN.
The hypotensive activity and side effects of methyldopa, clonidine and guanaficine. Hypertension. 1984; 6(5 Pt 2): II-28 – II-33. - Robertson D, Tung C-E, Goldberg MR, Hollister AS, Gerkens JF, Oates JA. Antihypertensive metabolites
of a-methyldopa. Hypertension. 1984; 6(5 Pt 2):
II-46 – II-50. - Gillespie L, Oates JA, Crout JR, Sjoerdsma A. Clinical and chemical studies with alpha-methyl-dopa in patients with hypertension. Circulation. 1962; 25:
281-291. - Tollefson S, Arentz-Hansen H, Fleckenstein B, et al. HLA-DQ2 and DQ8 signatures of gluten T-cell epitopes in celiac disease. J Clin Invest. 2006; 116(8): 2226-2236
- Jabri B, Sollid LM. T-cells in celiac disease. J Immunol. 2017; 198(8): 3005-3014.
Issue
Related topics
Assays, Drug Discovery, HTS (High Throughput Screening), Research & Development (R&D), t-cells