O-glycan analysis of therapeutic proteins enabled by O-glycoprotease
Posted: 29 June 2021 | Christopher H Taron (New England Biolabs [NEB]), Saulius Vainauskas (New England Biolabs [NEB]), Xiaofeng Shi (New England Biolabs [NEB]) | No comments yet
Glycosylation of therapeutic proteins is important to biologic drug development and is a critical quality attribute that is monitored during manufacturing. Analysis of O-glycans is technically challenging compared to that of N-glycans. In this review, Xiaofeng Shi, Saulius Vainauskas and Christopher H Taron summarise current O-glycan analytical approaches, describe the dearth of reliable tools for O-glycan analysis and highlight the benefits of a recent technological advancement in the sector.

Therapeutic proteins and O-glycosylation
Glycosylation is one of the most common and elaborate post-translational modifications. It profoundly affects biophysical, biochemical and, hence, biological properties of glycoproteins, including many protein therapeutics.1 Unlike linear nucleic acids or proteins, glycans are synthesised without templates and are often branched, resulting in tremendous degrees of structural heterogeneity. Specific changes in the glycan structures can alter the stability and efficacy of these biotherapeutics.2 As such, glycans of these drug molecules are extensively studied and closely monitored during drug discovery, development and manufacturing.
There are two major types of glycosylation on proteins: N-glycosylation, in which a paucimannose core structure is attached to the asparagine of a canonical N-X-S/T sequence (X ≠ P); and O-glycosylation, the focus of this review. The most prevalent form of O-glycosylation contains an N-acetylgalactosamine (GalNAc) attached to the hydroxyl group of a serine or threonine. Both the paucimannose core structure in N-glycans and the GalNAc in O-glycans (also called mucin-type) can be further extended with other monosaccharides and undergo post-glycosylation modifications.
Deviations in glycosylation patterns can significantly affect a therapeutic’s efficacy, stability and half-life in the blood stream”
Recent decades have seen continuous growth in the number of recombinant proteins used as therapeutics and, more recently, as vaccines.3 These proteins include enzymes for use in enzyme replacement therapy, hormones for a variety of indications, and well over 100 monoclonal antibodies (mAbs) and associated fusion proteins for the treatment of many diseases, including cancer and autoimmune disorders.4,5 Most of these proteins are recombinantly produced in mammalian cell lines and possess glycosylation patterns that reflect those of the host cells. Deviations in glycosylation patterns can significantly affect a therapeutic’s efficacy, stability and half-life in the blood stream. Glycan analysis is, therefore, woven into the entire process of development and manufacturing of therapeutic proteins, including cell line screening, process development, quality control and regulatory filing. Moreover, for biosimilars, matching glycosylation to that of the innovator drug is a foremost concern that precedes many other aspects of drug development.6
Of the two types of protein-bound glycans, N-glycans have, by far, received the most attention, with an abundance of literature, dedicated workflows, sample preparation kits and informatics tools. The International Conference on Harmonization (documents Q5E and Q6B) recommends that: “…the structure of the carbohydrate chains, the oligosaccharide pattern [antennary profile], and the glycosylation site[s] of the polypeptide chain is analysed, to the extent possible.”7,8 However, the US Pharmacopoeia only includes LC-FLD (MS) of N-glycan analysis in its Oligosaccharide Analysis Chapter.9
There is a clear need for better tools and methods to enable more reliable determination of O-glycan structure and O-glycan attachment sites”
The dearth of information on O-glycan analysis for biotherapeutics can be attributed to several factors. First, a large proportion of therapeutic proteins are mAbs. Most monoclonals possess only a single N-glycan on the Fc region of the heavy chain (eg, Asp297 for human IgG) and no O-glycans. Second, there is no consensus amino acid sequence for O-glycosylation, and site occupancy can be highly variable. Third, the most common form of O-glycosylation (mucin-type) tends to be highly clustered within proteins, which makes both enzymatic cleavage and structural analyses far more challenging. Lastly, there is no known broad specificity O-glycosidase that is able to release O-glycans for structural profiling. Chemical release methods exist, but these subject released glycans to degradation and information loss. Therefore, there is a clear need for better tools and methods to enable more reliable determination of O-glycan structure and O-glycan attachment sites.
Current O-glycan analysis approaches
A common method of glycan analysis involves the release of glycans from a glycoprotein prior to their structural profiling (Figure 1). This method is well suited for N-glycan analysis because the enzyme PNGase F can effectively release a broad range of N-glycans. Unfortunately, there is no analogous broad-specificity enzyme for O-glycan analysis. A few endo-α-N-acetylgalactosaminidases from microbial sources can remove Galβ1,3- GalNAcα (Core 1) or GlcNAcβ1,3-GalNAcα (Core 3) disaccharides from a serine or threonine.10,11 However, any further extension beyond the Core 1 or 3 structure (eg, sialic acid) will prevent the enzyme from cutting.
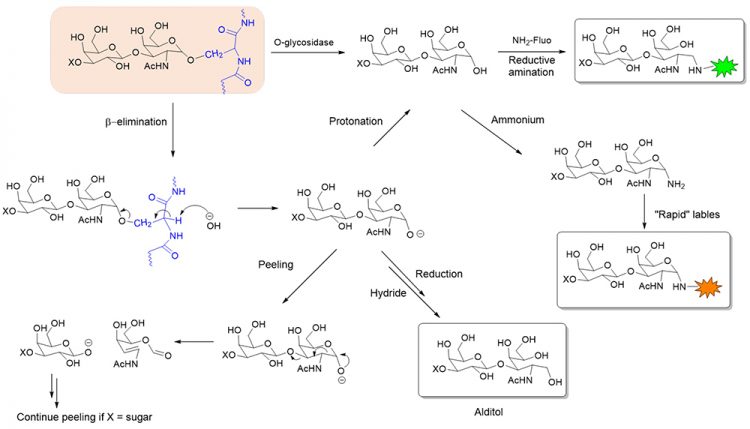

Figure 1: Chemical and enzymatic release and subsequent labelling of O-glycans. X = H or monosaccharide.
A more complete approach of releasing O-glycans utilises chemical deglycosylation in the presence of alkali (commonly termed β-elimination). This approach, however, suffers from several technical complications. First, β-elimination of the S/T-attached GalNAc that also has an immediate 1-3 linked monosaccharide (eg, Gal or GlcNAc) often causes a cascade of subsequent elimination reactions (termed ‘peeling reactions’), leading to degradation of the GalNAc. Second, a strong reductant (eg, NaBH4) is sometimes introduced to convert the reducing end of the released glycan to an alditol. This modification, while avoiding peeling, also prevents the glycan from being further derivatised with a fluorophore, or mass tag, to aid downstream analyses. A final drawback of this release approach, as with the enzymatic release, is that it provides no information on where the glycan was attached to the protein.
In contrast to the released glycan analyses, glycans can also be studied at the glycopeptide or glycoprotein level (Figure 2). Such analyses can yield information on glycosylation sites, glycan structure and the peptide backbone. This approach is especially useful for O-glycan analyses as O-glycosites are less predictable and site occupancy is often highly variable.
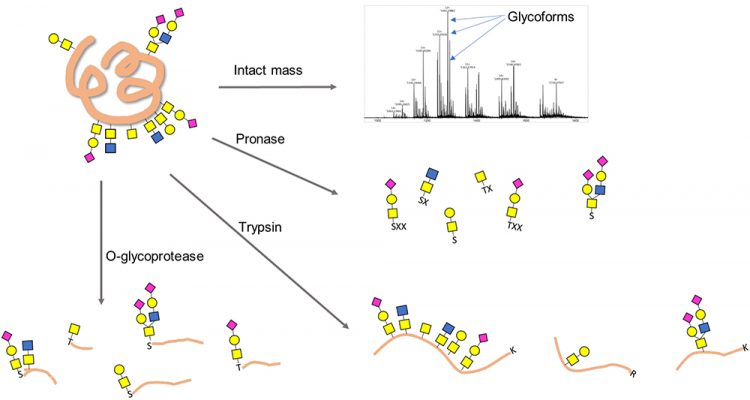

Figure 2: Schemes for O-glycoprotein and O-glycopeptide analysis. X denotes a nonspecific amino acid.
A top-down approach of intact glycoprotein can potentially provide a plethora of information on not only glycosylation, but also other PTMs or protein variants.12 This approach requires ultra-high performance mass spectrometers, such as Fourier Transform Ion Cyclotron Resonance, as well as specialty dissociation methods, such as electron transfer dissociation (ETD) or electron capture dissociation (ECD). The lack of access to these sophisticated instruments as well as the absence of general data interpretation software for top-down glycoproteomics limits its wide application in industry.
The bottom-up or “shotgun” approach digests a protein non-selectively into short oligopeptides with one to several amino acids. These short peptides are then subject to enrichment, derivatisation and analysis. While this approach can produce a complete glycan profile, information on specific glycosites and site occupancies is often lost. A frequently used broad-specificity protease for this approach is Pronase.13
An O-glycoprotease that has broad specificity and low bias towards peptide sequence is highly enabling for O-glycan analysis and O-glycoproteomics in biopharma”
Trypsin is a more specific protease that can be used to produce peptide mixtures containing O-glycopeptides analysed by mass spectrometry (MS) along with peptide mapping.14 The primary drawback of using trypsin or other sequence‑specific proteases for O-glycan analysis is that the cleavage sites do not pinpoint the location of the glycan within any given peptide. Moreover, O-glycan-containing peptides, if not flanked by a trypsin site (lysine or arginine) in the vicinity of each glycosite, can be physically long and, therefore, can behave unfavourably in chromatography and MS. Finally, the highly clustered and repetitive nature of mucin-type O-glycans makes deconvolution of structural information by mass matching and MS/ MS fragmentation even more challenging.
Glycopeptide analysis enabled by O-glycoprotease
Ideally, an O-glycan-specific protease that possesses the following properties can maximally facilitate O-glycopeptide analysis. First, highly specific protease should be able to cleave at serine or threonine containing O-glycan to produce glycopeptides that directly indicate O-glycosylation sites. Second, glycoprotease must possess a broad substrate specificity for different O-glycan structures from a single GalNAc residue to complex O-glycans containing sialic acids. Third, this protease should display minimal peptide sequence specificity relative to the amino acids immediately adjacent to the serine/threonine. This can ensure unbiased O-glycopeptide production, regardless of the surrounding peptide sequence context.
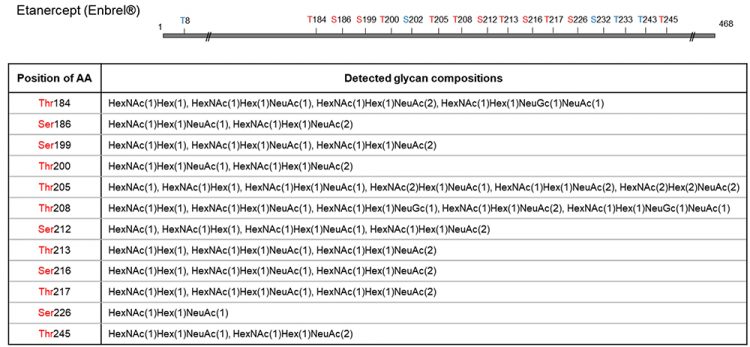

Figure 3: O-glycosite determination and O-glycan profiling of Etanercept using O-glycoprotease. Etanercept protein was digested with O-glycoprotease. The generated peptides were analysed with C18 column coupled with a QExactive Hybrid Quadrupole-Orbitrap mass spectrometer. MS/MS spectra were searched against selected protein and glycan databases using Byonic software. All glycopeptides were further validated using oxonium ions. Glycopeptide mapping detected 12 O-glycosites in total, with glycan compositions tabulated for each site, many of which heavily sialylated. Amino acids in blue are inferred glycosites from complementary peptides.
OpeRATOR (Genovis) is a protease that cuts strictly at the serine/threonine attached with a Gal‑GalNAc, but is unable to cleave or shows limited activity on common structures such as GalNAc-(Tn antigen) and sialylated O-glycan structures.15 In fact, sialidase is bundled in the kit and desialylation is a pre-requisite for broader peptide cleavages. While it is the first commercial enzyme to facilitate glycosite determination, its limitation in specificity makes it unsuitable for O-glycan profiling or O-glycan structural analysis.
New England Biolabs recently launched a new O-glycoprotease that meets all three criteria described above for maximally facilitating O-glycopeptide analysis. The enzyme cleaves the immediate N-terminal of O-glycosylated serine/threonine residue; it exhibits much broader substrate specificity, with its activity unaffected by sialic acids; and it displays low preference at P1 position (ie, N-terminal) of the serine/ threonine. Moreover, it can be used alone or in combination with another protease, depending on the O-glycosylation patterns, to produce both O-glycosite and O-glycan profiles. As an example, Etanercept (Enbrel), when incubated with O-glycoprotease, produced glycopeptides that indicates 12 O-glycosylation sites, as well as rich information in the O-glycan compositions in each site (Figure 3).
Conclusion
The ability to produce and characterise O-glycopeptides is vital for O-glycan analysis due to the low predictability of the O-glycosites and high degree of variability in O-glycan structure and occupancy. An O-glycoprotease that has broad specificity and low bias towards peptide sequence is highly enabling for O-glycan analysis and O-glycoproteomics in biopharma. The abundance of information generated from this approach coincides with the latest trend of Multiple-Attribute Measurement (MAM) in drug characterisation.16 O-glycoprotease not only greatly facilitates O-glycan analysis, but can also potentially be adopted in MAM experiments to characterise therapeutic glycoproteins.
About the authors


Xiaofeng Shi, PhD (The Ohio State University) is Development Group Leader at New England Biolabs (NEB), responsible for the applications and product development efforts in protein expression, analysis and glycobiology product portfolio.


Christopher H Taron, PhD (University of Illinois Urbana-Champaign) is the Scientific Director of Protein Expression and Modification research at NEB.


Saulius Vainauskas, PhD Saulius Vainauskas, PhD (Vilnius University, Lithuania) is a Researcher Scientist III at the same division.
Their research focuses on new enzymatic tools and analytical approaches for glycobiology and glycomics, as well as developing new technologies for heterologous protein production in various yeast hosts.
References
- Varki A, Cummings RD, Esko JD, et al. 2015. Editors. Essentials of glycobiology. 3rd ed, Cold Spring Harbor Laboratory Press.
- Zeerleder S, Engel R, Zhang T, et al. 2021. Improving in vivo clearance rate of highly glycosylated recombinant plasma proteins for therapeutic use. Pharmaceuticals. Jan 11;14(1):54.
- Solá RJ, Griebenow K. 2010. Glycosylation of therapeutic proteins: an effective strategy to optimize efficacy. BioDrugs. Feb 1;24(1):9-21.
- Strohl WR. Fusion proteins for half-life extension of biologics as a strategy to make biobetters. BioDrugs. 2015 Aug;29(4):215-39.
- Strohl WR. 2018. Current progress in innovative engineered antibodies. Protein Cell. Jan;9(1):86-120.
- Hajba L, Szekrényes Á, Borza B, Guttman A. 2018. On the glycosylation aspects of biosimilarity. Drug Discovery Today. Mar;23(3):616-625.
- ICH, Q5E Specifications: Test Procedures and Acceptance Criteria for Biotechnological/Biological Products, EMA Document CPMP/ICH/5721/03 (Geneva, 2003).
- ICH Q6B Specifications: Test Procedures and Acceptance Criteria for Biotechnological/Biological Products, EMA Document CPMP/ICH/365/96 (Geneva, 1999).
- USP40 NF35, Published General Chapter <212> Oligosaccharide Analysis, 2017
- Koutsioulis D, Landry D, Guthrie EP. 2008. Novel endo-alpha-N-acetylgalactosaminidases with broader substrate specificity. Glycobiology. Oct;18(10):799-805.
- D’Atri V, Nováková L, Fekete S, et al. 2019. Orthogonal middle-up approaches for characterization of the glycan heterogeneity of etanercept by hydrophilic interaction chromatography coupled to high-resolution mass spectrometry. Anal Chem. Jan 2;91(1):873-880.
- Yu Q, Wang B, Chen Z, et al. 2017. Electron-transfer/higher-energy collision dissociation (ETHCD)-enabled intact glycopeptide/glycoproteome characterization. J Am Soc Mass Spectrom. Sep;28(9):1751-1764.
- Stavenhagen K, Plomp R, Wuhrer M. 2015. Site-Specific Protein N- and O-glycosylation analysis by a C18-porous graphitized carbon-liquid chromatography-electrospray ionization mass spectrometry approach using pronase treated glycopeptides. Anal Chem. Dec 1;87(23):11691-9.
- Houel S, Hilliard M, Yu YQ, et al. 2014. N- and O-glycosylation analysis of etanercept using liquid chromatography and quadrupole time-of-flight mass spectrometry equipped with electron-transfer dissociation functionality. Anal Chem. Jan 7;86(1):576-84.
- Nordgren M, Nägeli A, Nyhlén H, Sjögren J. 2021. Mapping O-glycosylation sites using OpeRATOR and LC-MS. Methods Mol Biol. 2271:155-167.
- Rogers RS, Abernathy M, Richardson DD, et al. 2017. A view on the importance of “Multi-Attribute Method” for measuring purity of biopharmaceuticals and improving overall control strategy. AAPS J. Nov 30;20(1):7.
Issue
Related topics
Analytical techniques, Biopharmaceuticals, Drug Manufacturing, ICH guidelines, Proteins, QA/QC, Therapeutics
Related organisations
International Conference on Harmonization (ICH), The United States Pharmacopoeia (USP)