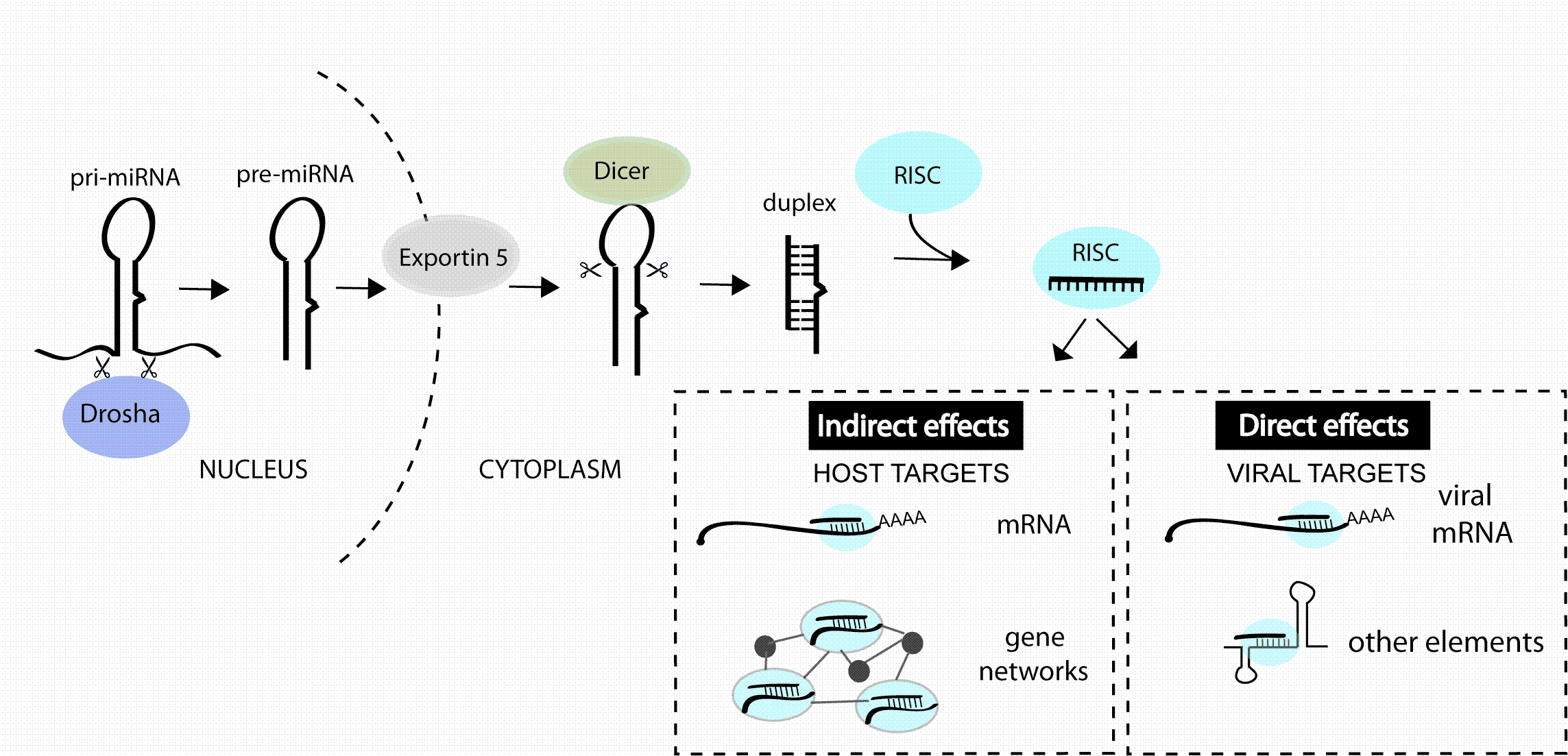
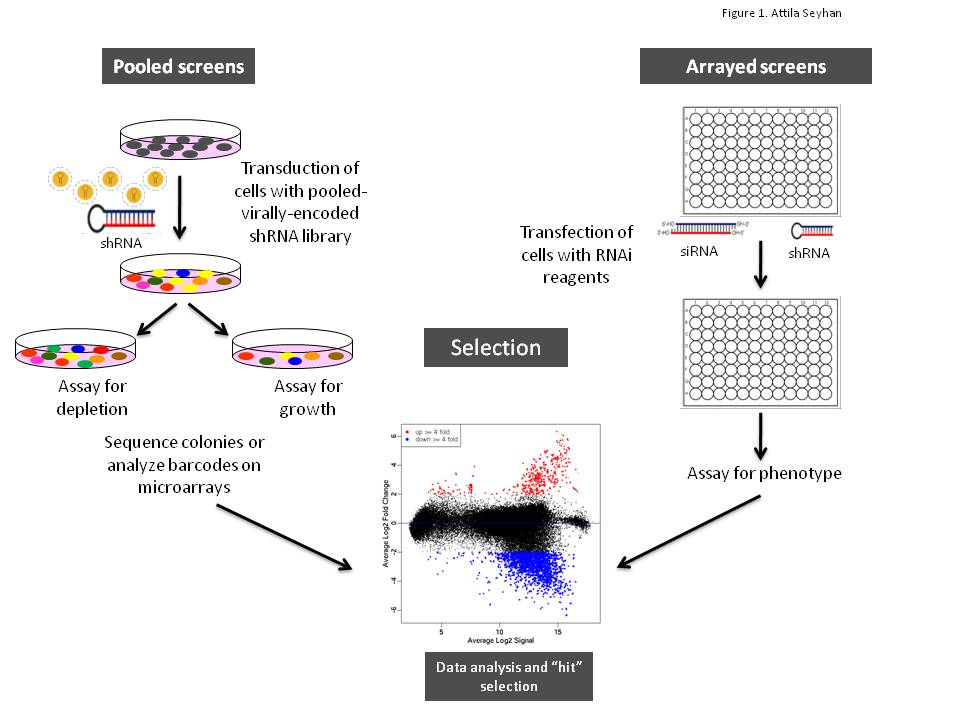
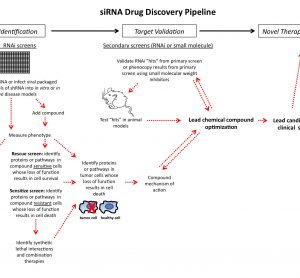
article19 October 2011 | By Marie Lundbæk, Department of Cancer Research and Molecular Medicine, Norwegian University of Science and Technology and Pål Sætrom, Department of Cancer Research and Molecular Medicine & Department of Computer and Information Science, Norwegian University of Science and Technology
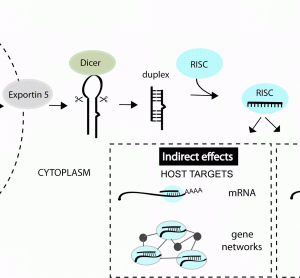
RNA interference (RNAi) is now a standard tool in molecular biology. Short interfering RNAs (siRNAs) for knocking down your favourite human gene are only a couple of mouse-clicks away at your favourite reagent supplier’s website. Moreover, in contrast to initial attempts at siRNA design, these siRNAs usually give potent target…