Optimising bispecific antibody clonal cell selection with high-throughput analytics
Posted: 22 December 2022 | Alice Harrison (Lonza), Stuart Jamieson (Lonza) | No comments yet
It is possible to manufacture monoclonal antibodies using traditional methods, but the challenges inherent in bispecific antibodies such as low titer, mismatched chains, unwanted fragments and higher aggregation levels require a heightened analytical focus on clonal cell selection. Here, Stuart Jamieson and Alice Harrison at Lonza discuss high-throughput analytical strategies that enable right-first-time selection of the optimal clone to take through to clinical manufacture.


Innovations in antibody design have led to an increasingly diverse range of therapeutic molecules in development including bispecific antibodies (bsAbs). Eight bispecific antibodies have been approved by the European Medicines Agency, including four novel products in 2022.1 Such rapid expansion provides opportunities for significant therapeutic advancement.
In contrast to natural antibodies, bsAbs are highly engineered therapeutic molecules designed to bind to two antigen epitopes simultaneously. Innovative bsAb designs, as highly targeted medicines, hold the promise of changing the way we treat cancer and other diseases.2 A bsAb can have clinical advantages over conventional monoclonal antibodies (mAbs) including targeting functionally interrelated checkpoint inhibitors, directing immune responders to cancer cells, enhancing specificity and decreasing off-target toxicity. While the first successful approvals of bsAbs underscore the potential of this class of antibodies, they also highlight the manufacturing challenge; the first bsAb, blinatumomab, took 13 years to advance through clinical development.3 As bsAbs are highly engineered and often comprise three or more polypeptide chains, they have increased production challenges compared to mAbs. These challenges include low product yields, erroneous chain assembly, high aggregate levels and adverse fragmentation.
As bsAbs are highly engineered and often comprise three or more polypeptide chains, they have increased production challenges compared to mAbs”
To achieve accelerated timelines and overcome bsAb manufacturing challenges, a focus on critical path activities is required.4 One of the first activities to fall onto the critical path, and potentially cause substantial drug development delays, is clone selection. Rapid development of clonal cell lines is required to facilitate material manufacture for non-clinical toxicological studies, and master cell bank (MCB) creation to enable clinical manufacture. Traditionally, preparing the MCB can be time consuming and expensive, taking six months or more. The success of the MCB choice impacts both regulatory approval prospects and the manufacturability of the bsAb. A delicate balance between the high-resource obligations of extensive testing and the importance of the choice means manufacturers are motivated to employ improved strategies for clone screening and characterisation at the earliest stages.
Rapid development of clonal cell lines
The development pathway for mAbs has been established over many years”
The aim of clonal bsAb cell line development is to identify a single clonal line that stably secretes gram-per-litre quantities of product with desired critical quality attributes (CQAs). For a bsAb these attributes include correctly assembled product in addition to minimal aggregation and fragmentation. While construct design optimisation and improved transfection technologies can partially overcome these challenges, as discussed in an earlier article, enhanced cell line development strategies and analytical screening improve expression profiles and yield of bsAbs.5
Cell line development workflows can be intricate and the development pathway for mAbs has been established over many years. Chinese hamster ovary (CHO) cells are the most commonly used mammalian expression hosts. Platforms for CHO cell line establishment are routinely optimised to facilitate robust, scalable production, which can be tailored to achieve clinically‑relevant post‑translational modifications. Once an expression construct is selected, cells are transfected, stable pools generated, clonal cells isolated and monoclonality is tested. After monoclonality confirmation, cells are expanded, productivity is assessed and clone stability is confirmed. This process is outlined in Figure 1.
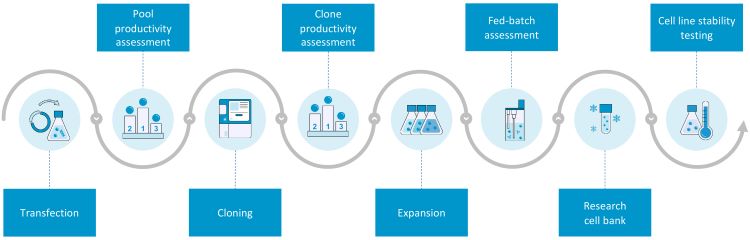

Figure 1: Overview of the cell line development process at Lonza.
In recent years significant improvements in automation of cell line development have reduced timelines for demonstrating productivity and clonality of lead cell lines from months to weeks. Technologies include advanced platforms like ClonePix® with monoclonality assurance, CellCelector®, Beacon® and Cyto-Mine®, which automate cloning, productivity screening and, in some cases, identify clones with increased aggregation.6 At Lonza the Beacon system is used to rapidly screen and isolate thousands of highly productive clones generated using Lonza’s GS Xceed® PiggyBac® expression system. During a study performed at Lonza, approximately 3,000 individual cells from a polyclonal pool were isolated into pens on an optofluidic chip. The pens were automatically screened for monoclonality and pens containing polyclonal populations were rejected. As the cells were developed, their productivity and growth characteristics were continually monitored. Using this data, it was possible to select a lead cell line that was scalable and produced 8 g/l across 15ml to 200l (see Figure 2).
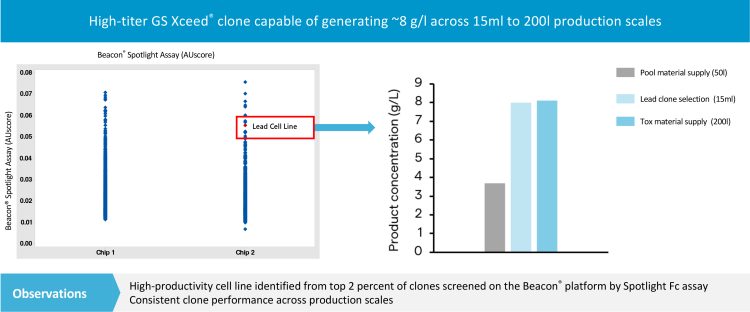

Figure 2: Assessment of thousands of individual clones using the Beacon Spotlight Assay for binding of the Fc antibody domain. Polyclonal pool material is sorted into monoclonal populations for selection of a lead clone and material supply for toxicological studies.
Product quality screening during clonal cell line selection
The increasing variety of bsAb formats under development leads to a propensity for undesirable common molecule characteristics to occur in varying levels across the clones. Therefore, it is necessary to evaluate CQAs during clone selection. It is beneficial to go beyond the standard analytical toolbox and perform additional testing. By conducting this analysis, it is possible to identify clones with an optimal balance of performance, ie those that are high yielding but also have good CQAs and low levels of product-related impurities. Selection of the clone with the best characteristics can reduce the purification challenge, potentially leading to more efficient production process and better overall yield compared to selecting on titer alone.
Selection of the clone with the best characteristics can reduce the purification challenge, potentially leading to more efficient production process and better overall yield compared to selecting on titer alone”
While it is possible to perform certain analysis, as discussed below, without the need to purify samples, other methods require partial purification of the clonal supernatants. This purification needs to primarily reduce the level of host cell impurities and retain the product-related ones in order to allow a comparison of the clones and the product CQAs. To facilitate the selection of high-performing clonal cell lines, a rapid purification method that supports analysis of a large number of cell line samples is required. Fortunately, high-throughput automated liquid handling purification platforms can be utilised. These platforms employ affinity chromatography to capture and collect the product and product-related species for further analysis.
Rapid analytical strategies for clone assessment
The most advanced cloning and screening platforms provide initial screening capabilities and estimates of productivity. However, they are not, for the most part, capable of assessing highly relevant CQAs for bsAbs, such as chain pairing, fragmentation and aggregation.
Analytical testing of bsAbs is more challenging than standard mAbs due to the requirement for product-specific methods to be employed. The need for high-throughput (HT) analytics to address these challenges is clear. This need has led to advancements in HT mass spectrometry (MS) techniques, including:
- affinity-native protein A-MS for assessment of chain pairing and fragmentation without purification7
- competitive binding-MS for analysis of multiple PQAs and rapid HT N-glycan analysis using combined LC/MS
- capillary electrophoresis (CE) for HT glycan screening of clones.8
Rapid establishment of toolbox methods for chain mispairing, such as Lonza’s RP-HPLC MS chain pairing technology,4 can provide key insights into the quality of bispecific assembly at a critical stage of clonal selection, while minimising impact upon project timeline.
Rapid establishment of toolbox methods for chain mispairing, such as Lonza’s RP-HPLC MS chain pairing technology,4 can provide key insights into the quality of bispecific assembly at a critical stage of clonal selection, while minimising impact upon project timeline.”
A key functional advantage of bsAbs over mAbs is the ability to bind two separate antigens. It is critical to establish dual specificity, and in doing so confirm correct assembly, early in the cell development process. A barrier to this has been the requirement for low-throughput binding and activity assays, such as enzyme-linked immunosorbent assays (ELISAs) and cell-based potency assessments. However, recent advances in biolayer interferometry (BLI) and surface plasmon resonance (SPR) sensor systems have overcome these barriers.9 An HT method was developed at Lonza for assessment of chain pairing and bsAb functionality; the binding of the antigens demonstrates whether each bsAb functional domain is active, while the ratio of the binding relates to the chain assembly. This technology enables ranking of clones based on correct molecular assembly. Including a control antibody for calibration curve generation and using an HTP surface plasmon resonance instrument, allows information on binding quality, titer and chain pairing of potentially hundreds of clones within a single automated experiment. The use of pre-established technologies, such as a CM5 protein A sensor surface, permits rapid method establishment and a minimal requirement for assay optimisation. An SPR study performed at Lonza in which bsAb clones were selected for further assessment, demonstrated the method enables chain pairing assessment, shown in Figure 3, where clones that yield antibodies with binding ratios close to one were correctly assembled. This allows rapid information generation to select the most suitable candidate as a lead clone.
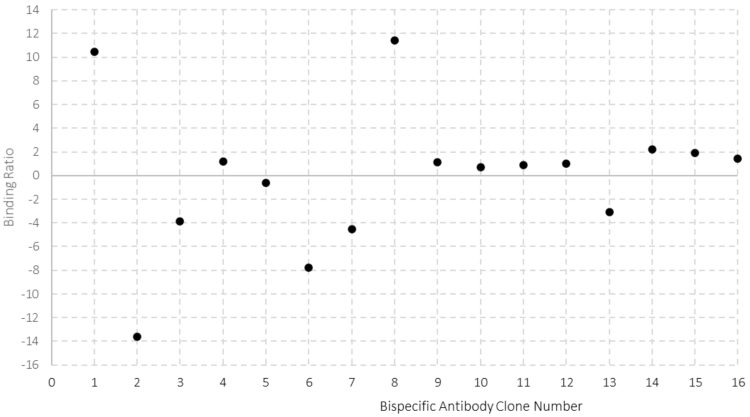

Figure 3: Assessment of bsAb chain assembly using surface plasmon resonance. An assay was developed to establish assembly of Fc containing bispecific molecules. Binding levels are monitored and the ratio between the binding of antigen 1 and antigen 2 indicates incorrectly assembled molecules, where a ratio of 1 indicates a correctly assembled heterodimer.
Further work in this area could enable not only a qualitative binding assessment and quantitative titer and chain pairing ranking, but also a detailed kinetic characterisation and potency determination of large numbers of bispecific clones in a single experiment.
Future perspectives on cell line selection
Rapid selection of a single master cell line expressing more complex antibody molecules from thousands of possible clones lends itself to the application of machine learning tools and software-assisted cell line selection”
Speed, performance and quality are all important drivers for a bsAb cell line development strategy to ensure that a high value clone is selected within an accelerated timeline and to reduce the manufacturing challenge. In recent years, enabling technologies have led to a large reduction in workflows as well as incorporation of HT analytics to help make informed selection decisions. This is helping us move towards a fully automated workflow, including product relevant HT analytics and integrated data analysis. In addition, the challenge of rapid selection of a single master cell line expressing more complex antibody molecules, such as bsAbs, from thousands of possible clones lends itself to the application of machine learning tools and software-assisted cell line selection. There is a growing move away from established, conventional cell culture and analysis techniques towards more digitally immersive cell development platforms. Future platforms will be built upon laboratory implementation of nanofluidic technology, for single cell level data collection, and cell line selection trough the predictive capabilities of machine-learning algorithms. If utilised appropriately such platforms will revolutionise clone selection and the creation of the MCB for bispecific and complex protein products, delivering increased efficiency. This will ultimately propel improved performance via precise decision making and enhanced control of quality attributes.
About the author


Stuart Jamieson is a Global Technical & CMC Director for Downstream Development at Lonza in Slough, UK. He has over 20 years’ experience in protein purification and analysis, process development, scaleup and clinical manufacture for early- to late-phase products gained working with Lonza, CPI Biologics and another major CDMO. Stuart’s expertise spans providing technical leadership for a diverse range of product types and is underpinned by a degree in biochemistry and a PhD in biochemistry and structural biology from The University of Sheffield, UK.


Alice Harrison is the Global Technical Director (CMC & Analytics) at Lonza, in Slough, UK and joined the company in 2022. Prior to that Alice worked within the biopharmaceutical industry for over 20 years, where she founded and led businesses supporting the development of antibody and complex protein therapeutics, gaining experience in large molecule analysis and an in-depth understanding of the bioanalytical challenges of complex molecule programmes. Her experience is supported by a degree in chemistry from Queen Mary College (University of London, UK) and a DPhil in molecular immunology from the University of Oxford, UK.
References
- The Antibody Society. Therapeutic monoclonal antibodies approved or in regulatory review. [Internet] Available at: www.antibodysociety.org/antibody-therapeutics-product-data [Cited: Oct2022].
- Lim SM, Pyo KH, Soo RA, Cho BC. The promise of bispecific antibodies: Clinical applications and challenges. Cancer Treat Rev. 2021 Sep;99:102240. Available at: doi.org/10.1016/j.ctrv.2021.102240
- Friberg G, Reese D. Blinatumomab (Blincyto): lessons learned from the bispecific t-cell engager (BiTE) in acute lymphocytic leukemia (ALL). Ann Oncol. 2017 Aug 1;28(8):2009-2012. Available from: doi:10.1093/annonc/mdx150.
- Jamieson S, Harrison A. Accelerating timelines for development and manufacture of multi-specific antibodies. European Pharmaceutical Review 2022;27(04):48.
- Jamieson S, Harrison A. Rapid method development to overcome challenges of bi-specific antibody purification. European Pharmaceutical Review 2022; 27 (03);6.
- Tejwani V, Chaudhari M, Rai T, Sharfstein ST. High‐throughput and automation advances for accelerating single‐cell cloning, monoclonality and early phase clone screening steps in mammalian cell line development for biologics production. Biotechnology Progress. 2021;37(6).
- Sawyer WS, Srikumar N, Carver J, et al. High-throughput antibody screening from complex matrices using intact protein electrospray mass spectrometry. Proceedings of the National Academy of Sciences. 2020;117(18):9851–6.
- Goel M, Srivastava R, Gupta S, et al. 2N-Glycan Analysis: Rapid preparation and screening of biosimilar candidates by LC/MS and CE. Application Note: 5994-0943EN.pdf (agilent.com)
- Li H, Kumaraswamy S, Rubio-Marrero E, et al. Application Note: A rapid method to quantitatively screen bispecific antibodies using Protein A and Octet® His1K Biosensors. sartorius.com.
- X Tao W, Ahmed W, Guo M, et al. Selection of high-producing clones by a relative titer predictive model using image analysis. Ann Transl Med. 2021 Jul;9(14):1144. Available at: doi: 10.21037/atm-21-2822.
This article featured in European Pharmaceutical Review Issue 6 2022. Sign up for free to receive future issues!
Issue
Related topics
Analytical techniques, Antibodies, Biopharmaceuticals, Cell culture automation, Manufacturing, Production, Technology