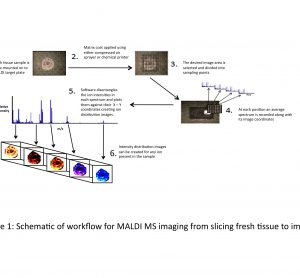
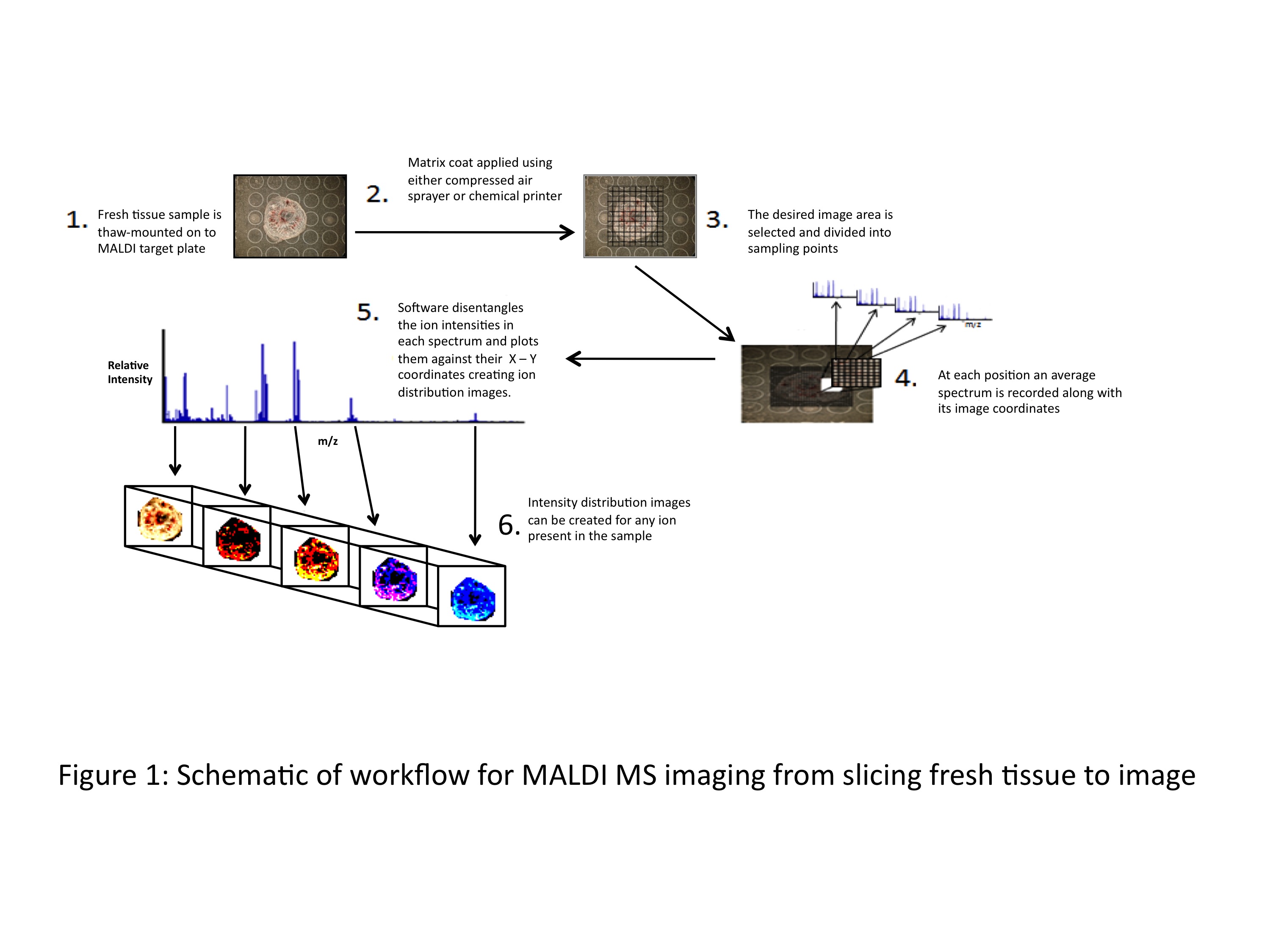
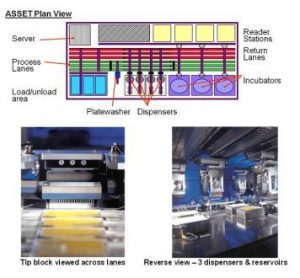
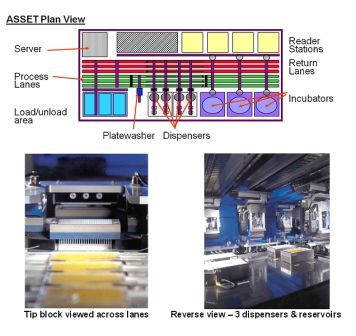
article29 October 2010 | By Brian Flatley
Dept of Chemistry, University of Reading, Reading and Harold Hopkins Dept of Urology, Royal Berkshire NHS Foundation Trust Hospital, Reading and Peter Malone
Harold Hopkins Dept of Urology, Royal Berkshire NHS
Foundation Trust Hospital, Reading and Rainer Cramer
Dept of Chemistry, University of Reading, Reading
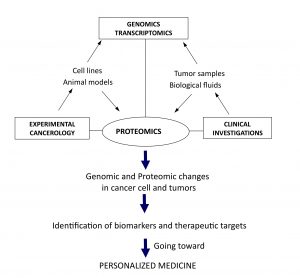
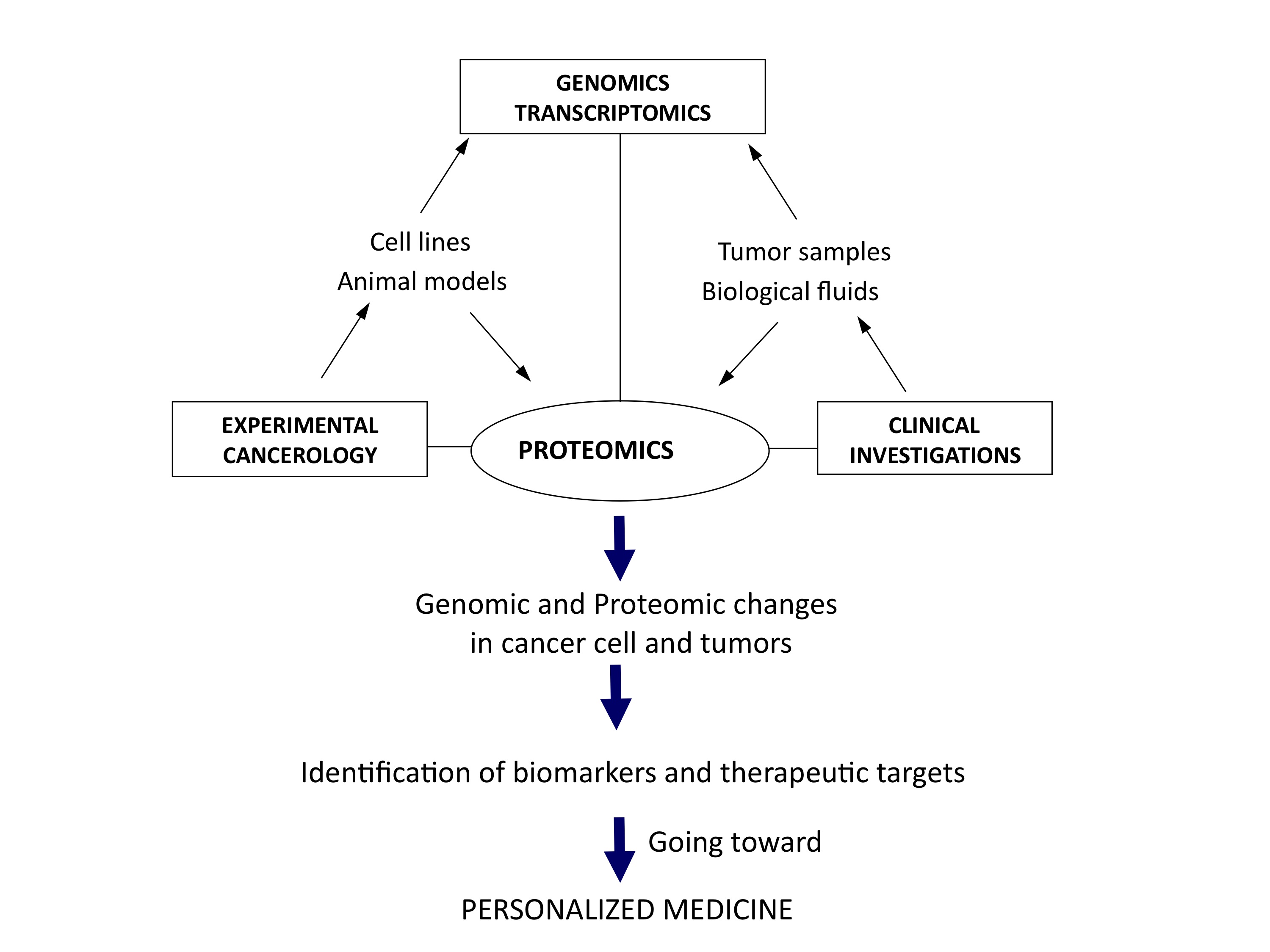
Each year, approximately 10,000 men in the UK die as a result of prostate cancer (PCa) making it the third most common cancer behind lung and breast cancer. Worldwide, more than 670,000 men are diagnosed every year with the disease. Current methods of diagnosis of PCa mainly rely on the…