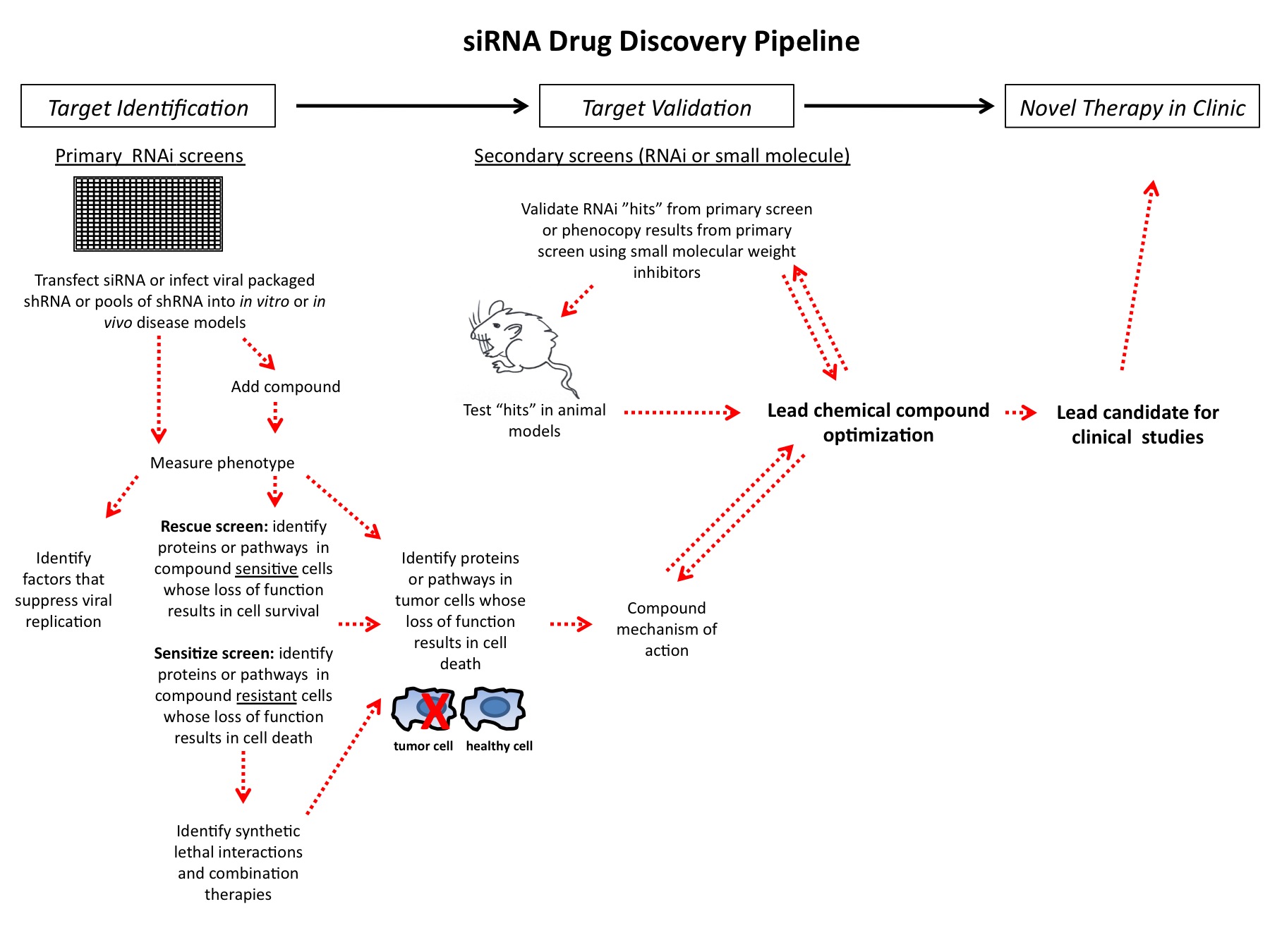
Many hands make light work: why the new Bespoke Gene Therapy Consortium is a godsend for ambitious biopharma teams
In late October, we learned that an exciting and ambitious new entity – the Bespoke Gene Therapy Consortium (BGTC) – had come into being in the US, designed to encourage the delivery of more gene therapies for rare diseases. The consortium is an interesting construct, headed and funded by the…