Advanced MALDI imaging breaks ground in pharmaceutical research
Posted: 3 September 2019 | Dr Carsten Hopf (Mannheim Technical University), Dr Rohan Thakur (Bruker Daltonics) | No comments yet
Dr Carsten Hopf and his team are using MALDI imaging in several applications, including drug metabolite and formulation distributions in tissues. This article investigates how this content-rich technique is enabling researchers to gain unique insights to facilitate faster and better drug discovery.

MATRIX-ASSISTED laser desorption/ionisation (MALDI) imaging is a non-radioactive, label-free and non-destructive technique that rapidly localises biomolecules – from metabolites to proteins – without prior knowledge of their presence.1 A single MALDI imaging measurement produces up to several thousand distribution maps, or ion images, to reveal greater insight and understanding of molecular makeup and regional heterogeneity.
Application areas for MALDI imaging are diverse and growing, driven by the label-free nature of the technique and the ability to differentiate compounds by molecular mass. Untargeted discovery studies, such as those in clinical research, seek to uncover novel biomarker signatures of disease and treatment. MALDI imaging is also revolutionising pre-clinical drug discovery pipelines by providing direct distribution monitoring of therapeutic compounds, their metabolites and target engagement.
MALDI imaging in pharma
Drug, drug metabolite and drug formulation distribution
At the Center for Mass Spectrometry and Optical Spectroscopy (CeMOS) at Mannheim Technical University, a broad range of mass spectrometry (MS) instruments are used to aid investigations into emerging applications in the pharma industry. This includes exploring typical applications for MALDI imaging in pharma, which, among other things, involves the distribution of drugs and their metabolites in tissues. The molecular composition of every drug is transformed in the liver, which leads to a change in mass that can be observed in the same dataset. The distribution of the parent drug can then be compared to its metabolites.
For example, the laboratory uses this approach to determine the co-localisation of drugs and drug metabolites with tumours, based on tumour-selective lipids that indicate its location.
Carrier particles, such as lipid nanoparticles, can change the drug half-life or distribution in the body, so everything relating to the drug should be imaged”
Recently, MALDI imaging was used to confirm the rapid in situ distribution of Fasudil – a drug approved for cerebrovascular bleeding – and its metabolite Hydroxyfasudil, in the liver and stomach tissue of wild type (WT) and gastric tumour tissue of transgenic mice.2 Fasudil is an inhibitor of RHO-associated protein kinases (ROCK1/2), drug targets downstream of the RHOA GTPase. Gain-of-function mutations of the latter had been implicated in diffuse-type gastric carcinoma,3 but the effectors were unknown. Data from this study suggests that ROCK1/2 may be suitable targets as a novel strategy for treating gastric cancer (GC), but the limited distribution of Fasudil into tumour tissue indicates the need for ROCK inhibitors optimised for tumour penetration. The identity of potentially translatable candidate lipid biomarkers for GC was established using MALDI magnetic resonance MS (MRMS) and MALDI time-of-flight (TOF) MS.
Subsequent statistical analysis can be used to evaluate a drug’s tumour-targeting potential, which would be completely impossible without the high resolving power of MALDI imaging. For example, this is particularly important for inhaled drugs, which should be designed to not enter the systemic circulation, where they often have the potential to cause harm. MALDI imaging helps to identify whether the drug stays local to the lung.
The CeMOS institute is also interested in imaging drug formulations, because there are other elements, in addition to the active pharmaceutical ingredient (API), that combine to make a drug. Carrier particles, such as lipid nanoparticles, can change the drug half-life or distribution in the body, so everything relating to the drug should be imaged. The group has used MALDI imaging to visualise the locations of intact liposomal drug carriers for the simultaneous imaging of drugs and formulations in tissues.4 Indocyanine green (ICG) was used as cargo and two liposomal markers, 1,2-dipalmitoylsn- glycero‑3‑phosphoglycerol (DPPG) and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine, conjugated with monodisperse polyethylene glycol (PEG36-DSPE), were incorporated into liposomal carriers and administered to mice (Figure 1). Newly developed MALDI imaging instrumentation has high imaging speeds, which displays improvements in lipid analysis of whole organ sections and so allowed the group to visualise liposome integrity and distribution in mouse organs.
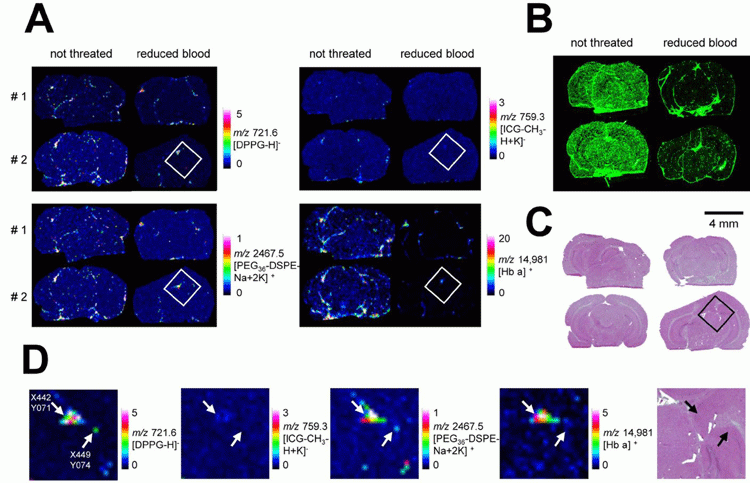

Figure 1: Images of brains of mice that were dosed with liposomes and localisation of the detected liposomes. A) MALDI MSI images were performed on brain slices of mice that were dosed with liposomes. Half of the mice were perfused before being sacrificed (right panels) to reduce the remaining blood in the tissue. MALDI images of 1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol (DPPG) and indocyanine green (ICG) were acquired in reflector negative ion mode, of monodisperse polyethylene glycol (PEG36-DSPE) in reflector positive mode and of HB α chain in linear positive mode. B) Fluorescence images performed on brain slices of mice that were dosed with liposomes. C) HE staining of tissue slices measured with MALDI MSI. D) Magnification of MALDI MS images of the boxed parts marked in perfused brain in A) and HE stained brain in C). Pixels indicated by an arrow shows the co-localisation of the liposomal components and haemoglobin at pixel X442 Y071 and the absence of HB at pixel X449 Y074. (Reproduced with permission from reference 4).
Drug response, pharmacodynamic biomarkers and drug formulations
The laboratory has also pioneered some additional MALDI imaging approaches. For example, the team was the first to image drug responses to elucidate the target engagement effects of drugs on tissues based on defined pharmacodynamic markers. In some cases, the marker of drug response might be known and can be imaged. If there is a defined protein and the drug action changes its mass, then the altered protein can be imaged. This is an indication not only of where the drug is located, but where exactly it works. Histone deacetylase inhibitors (HDACi) lead to mass changes characteristic of histone acetylation, which allow for quantitative label-free in situ monitoring of drug target engagement by MALDI-TOF MS in cancer cells.5
Multimodal imaging focusing on defined tissue morphologies
…the laboratory uses this approach to determine the co-localisation of drugs and drug metabolites with tumours”
Dr Hopf’s laboratory has also combined MALDI imaging with spectroscopic (infrared [IR]) imaging, a relatively fast technique that does not require tissue pre-processing, but which is not as content-rich as MALDI MRMS imaging.6 A computer processes the IR image and teases out specific areas of interest. MALDI imaging is then used to image these precise tissue regions; for example, those involved in learning and memory in the brain for a neurodegeneration project, to acquire data in those sections. This method combines the speed and ease of IR imaging with the precision and specificity of MALDI imaging.
Histopathology in pharmaceutical R&D
Histopathology-directed imaging is a new trend in MS imaging for pharma companies. It enables the user to obtain a molecular snapshot of the tissue area annotated by the histopathologist as indicating a pathological morphology. MALDI imaging generates an unbiased dataset for the analyst to determine whether it is the drug itself that is causing the abnormality, a changed version of the drug that was not anticipated or another factor not related to the drug at all. Using this technology, scientists can not only image expected results, such as a drug of known mass, but unknown entities as well. Thousands of masses can be measured in one dataset to produce new findings. For example, this can elucidate a drug’s possible toxic side effects to rule these out early in drug discovery before they make it to clinical trials, saving time and money.


Future work
It is likely that, given the growing interest in MALDI imaging for pharmaceutical research, instruments with higher sensitivity and better spatial resolution will emerge. New methods of obtaining high specificity to guarantee the visualisation of drug distribution at faster speeds would also greatly benefit future work.
More applications in pharma drug research will develop in line with technological advancements. For example, companies will not only use MALDI imaging for visualising drug distribution, but also drug action, histopathology and distribution of drug formulation ingredients. A larger number of use cases will be discovered in the coming years. For example, aside from MALDI imaging for pharma, the laboratory has begun investigating the use of quadrupole time-of-flight (QTOF) and MALDI-TOF MS for biopharmaceutical analysis. The group is interested in obtaining as much sequence information as possible and finding precise mass information on entire antibodies, antibody drug conjugates (ADCs) or fragments of antibodies (~25kDa). Biopharmaceutical development is a key area of interest for the laboratory’s future work, including monoclonal antibody (mAb) characterisation.
About the authors




References
- Schulz S, Becker M, Groseclose MR, Schadt S and Hopf C. Advanced MALDI mass spectrometry imaging in pharmaceutical research and drug development. Current Opinion in Biotechnology 2018; 55:51-59, https://doi.org/10.1016/j.copbio.2018.08.003.
- Hinsenkamp I, Schulz S, Roscher M, Suhr AM, Meyer B, Munteanu B, Fuchser J, Schönberg S, Ebert MPA, Wängler B, Hopf C, Burgermeister E. Rho-associated kinase 1/2-inhibitor fasudil attenuates tumor growth in a mouse model of diffuse-type gastric cancer. Neoplasia http://dx.doi.org/10.1016/j.neo.2016.07.002
- Kakiuchi M, Nishizawa T, Ueda H, Gotoh K, Tanaka A, Hayashi A, Yamamoto S, Tatsuno K, Katoh H, Watanabe Y, Ichimura T, Ushiku T, Funahashi S, Tateishi K, Wada I, Shimizu N, Nomura S, Koike K, Seto Y, Fukayama M, Aburatani H, Ishikawa S. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet 2014; 46(6):583-587. doi: 10.1038/ng.2984.
- Fülöp A, Sammour DA, Erich K, von Gerichten J, van Hoogevest P, Sandhoff R and Hopf C Molecular imaging of brain localization of liposomes in mice using MALDI mass spectrometry, Scientific Reports 2016;6, doi: 10.1038/srep33791.
- Munteanu B, Meyer B, von Reitzenstein C, Burgermeister E, Bog S, Pahl A, Ebert MP and Hopf C. Label-Free in Situ Monitoring of Histone Deacetylase Drug Target Engagement by Matrix-Assisted Laser Desorption Ionization-Mass Spectrometry Biotyping and Imaging. Analytical Chemistry 2014; 86(10), 4642−4647.
- Rabe JH, Sammour DA, Schulz S, Munteanu B, Ott M, Ochs K, Hohenberger P, Marx A, Platten M, Opitz CA, Ory DS and Hopf C. Fourier Transform Infrared Microscopy Enables Guidance of Automated Mass Spectrometry Imaging to Predefined Tissue Morphology. Scientific Reports 2018; 8:313.
Issue
Related topics
Biomarkers, Drug Discovery, Formulation, Imaging, Mass Spectrometry, Research & Development (R&D)
Related organisations
Center for Mass Spectrometry and Optical Spectroscopy (CeMOS), Mannheim Technical University